„Szerkesztő:Hollófernyiges/próbalap2” változatai közötti eltérés
Nincs szerkesztési összefoglaló |
Nincs szerkesztési összefoglaló |
||
| 1. sor: | 1. sor: | ||
{{Taxobox |
|||
{{Személy infobox |
|||
| image = Novel Coronavirus SARS-CoV-2.jpg |
|||
|név= John Howard Northrop |
|||
| image_caption = A SARS-CoV-2 transzmissziós elektronmikroszkópos képe |
|||
|kép= John Howard Northrop.jpg |
|||
| virus_group = iv |
|||
|képméret= |
|||
| ordo = ''[[Nidovirales]]'' |
|||
|képaláírás= |
|||
| familia = ''[[Coronaviridae]]'' |
|||
|születési név= |
|||
| subfamilia = ''[[Coronavirinae]]'' |
|||
|születési hely= [[Yonkers]] |
|||
| genus = ''[[Betacoronavirus]]'' |
|||
|születési dátum=[[1891]]. [[július 5.]] |
|||
| subgenus = ''[[Sarbecovirus]]'' |
|||
|halál helye=[[Wickenburg]] |
|||
| species = SARSr-CoV |
|||
|halál dátuma= [[1987]]. [[május 27.]]<br>(95 évesen) |
|||
| subspecies = SARS-CoV-2 |
|||
|halál oka = - |
|||
|sírhely = - |
|||
|állampolgárság = [[Amerikai Egyesült Államok|Egyesült Államok]] |
|||
|nemzetiség = |
|||
|házastárs= |
|||
|élettárs= |
|||
|szülei = - |
|||
|szakma= [[Biokémia|biokémikus]] |
|||
|iskolái= - |
|||
|művésznév= |
|||
|becenév= |
|||
|munkái= |
|||
|tisztség = - |
|||
|kitüntetései=[[kémiai Nobel-díj]] (1946) |
|||
|aláírás= |
|||
|weboldala= |
|||
|blog= |
|||
|IMDb= |
|||
|PORT.hu= |
|||
}} |
}} |
||
'''John Howard Northrop''' ([[Yonkers]], [[1891]]. [[július 5.]] – [[Wickenburg]], [[1987]]. [[május 27.]]) amerikai biokémikus. 1946-ban elnyerte a [[kémiai Nobel-díj]]at, mert kidolgozta több [[enzim]] és [[vírus]] kristályosítási eljárását. |
|||
A '''SARS-CoV-2''' (az angol ''Severe acute respiratory syndrome coronavirus 2'' rövidítése; magyarul ''súlyos akut légzőszervi szindróma-koronavírus 2'') a ''Coronaviridae'' családba tartozó, embereket fertőző vírustörzs, amely a 2019-es koronavírus-betegség (COVID–19) kórokozója. |
|||
==Tanulmányai== |
|||
John Howard Northrop 1891. július 5-én született a New York állambeli Yonkersben, John Isaiap Northrop és Alice Rich Northrop gyermekeként. Nagy múltú jenki családból származott, egyik őse még 1630-ban érkezett Amerikába és felmenői között előfordult a Princeton igazgatója vagy cukorgyártó iparmágnás. Apja a Columbia Egyetem zoológiai tanszékén tanított, de John születése előtt két héttel a zoológiai múzeumban kitörő tűzben az életét vesztette. A New York-i Normal College-ben botanikát oktató anyja egyedül nevelte fel őt. Jóval később, 1922-ben anyja is baleset áldozatává vált, amikor autója vonattal ütközött. |
|||
SARS-CoV-2 a Baltimore-féle osztályozási rendszerben a IV. csoportba (egyszálú, pozitív-szenz RNS-genommal rendelkező vírusok) tartozik, virionját lipidburok veszi körbe.<ref>{{cite journal |last1=Baltimore |first1=D |title=Expression of animal virus genomes. |journal=Bacteriological Reviews |date=1971 |volume=35 |issue=3 |pages=235–241 |doi=10.1128/MMBR.35.3.235-241.1971 |pmid=4329869 |doi-access=free }}</ref><ref name="gisaid" /> Taxonómiai szempontból a SARSr-CoV (súlyos akut légzőszervi szindrómához kapcsolódó koronavírus) faj egyik törzse,<ref name="CoronavirusStudyGroup" /> akárcsak közeli rokona, a 2002-2004-es SARS világjárványt okozó SARS-CoV-1.<ref name="NEJM-Stability"/><ref name="nihSARSr-CoV" /> |
|||
Northrop Yonkersben végezte el az elemi és középiskolát, majd a Columbia Egyetemen tanult tovább, ahol tagja volt a lövész- és vívóklubnak |
|||
A vírus zoonotikus eredetű, genetikai vizsgálatok szerint legközelebbi rokonai a denevérekben élnek.<ref name="NatureZhou" /><ref name="LancetNowcasting" /><ref name="MedVirEvolution" /><ref name="Proximal" /> Egyes feltételezések szerint a tobzoskák köztesgazdaként szolgálhattak a denevérek és az emberek között, de ez az elmélet még nincs bizonyítva.<ref name="WHO-SR22" /><ref name="DWPangolins" /> A vírus genetikai diverzitása alacsony, vagyis az emberre való "átugrása" nemrég, feltehetően 2019 végén következhetett be.<ref name="early" /> |
|||
Az epidemiológiai vizsgálatok szerint a vírus a védekező intézkedéseket nem hozó, immunológiailag nem védett közösségekben igen gyorsan terjed, egy beteg 1,4-3,9 másik embernek adja tovább a fertőzést. Terjedését a testi érintkezés vagy a köhögés, tüsszentés vagy akár a beszéd által generált cseppfertőzés biztosítja.<ref name="WHO2020QA" /><ref name="CDCTrans" /> Gazdasejtjébe az angiotenzin-konvertáló enzim-2 (ACE2) receptorhoz kapcsolódva jut be..<ref name="NatureZhou" /><ref name="NatMicLetko" /><ref name="HoffmanCell" /><ref name="NG-20200415" /> |
|||
===Terjedése=== |
|||
[[1915]]-ben a [[Columbia Egyetem]]en doktorált. [[1917]]-től [[1961]]-ig a [[Rockefeller Egyetem|Rockefeller Intézet]]ben dolgozott. A 30-as évek elején kutató munkájával igazolta és megerősítette [[James B. Sumner]] korábbi eredményeit, hogy az [[enzim]]ek kristályosíthatók. |
|||
A SARS-CoV-2 emberről emberre való terjedését már a vuhani járvány elején, 2020. január 20-án igazolták.<ref name="Chan24Jan2020" /><ref name="LiMar2020" /><ref name="Kessler17Apr2020" /><ref name="Kuo21Jan2020" /> Eleinte úgy vélték, hogy a kórokozó elsősorban a köhögés és tüsszögés kiváltotta cseppfertőzéssel terjed 1,5-2 méteren belül.<ref name="CDCTrans" /><ref name="NBCSpread" /> Lézeres fényszóródási vizsgálatokkal azonban kimutatták, hogy a közönséges beszéd is generál apró folyadékcsöppeket, amelyekben a vírus megbújhat;<ref>{{cite journal | vauthors = Anfinrud P, Stadnytskyi V, Bax CE, Bax A | title = Visualizing Speech-Generated Oral Fluid Droplets with Laser Light Scattering | journal = The New England Journal of Medicine | volume = 382 | issue = 21 | pages = 2061–2063 | date = May 2020 | pmid = 32294341 | pmc = 7179962 | doi = 10.1056/NEJMc2007800 }}</ref><ref>{{cite journal | vauthors = Stadnytskyi V, Bax CE, Bax A, Anfinrud P | title = The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 117 | issue = 22 | pages = 11875–11877 | date = June 2020 | pmid = 32404416 | pmc = 7275719 | doi = 10.1073/pnas.2006874117 }}</ref> sőt a vírusrészecskék magukban is kikerülhetnek a levegőbe.<ref name="NYT-20200704am">{{cite news |last=Apoorva Mandavilli |first=<nowiki>Apoorva]]</nowiki> |title=239 Experts With One Big Claim: The Coronavirus Is Airborne - The W.H.O. has resisted mounting evidence that viral particles floating indoors are infectious, some scientists say. The agency maintains the research is still inconclusive. |url=https://www.nytimes.com/2020/07/04/health/239-experts-with-one-big-claim-the-coronavirus-is-airborne.html |date=4 July 2020 |work=[[The New York Times]] |accessdate=5 July 2020 }}</ref> |
|||
A leülepedett cseppekkel fertőzött felületek fizikai érintése is veszélyes lehet.<ref name="WHO-Workplace" /> A kutatások szerint a SARS-CoV-2 műanyag- és acélfelületeken akár három napig, kartonpapíron egy napig, rézfelületen pedig négy óráig marad életképes.<ref name="NEJM-Stability" /> A detergensekkel (mint a szappan) való érintkezés felbontja a vírus külső lipidburkát és inaktiválja azt.<ref name="AtlanticSuccess" /><ref name="NatGeoSoap" /> A vírus RNS-ét kimutatták a beteg különféle testfolyadékaiból, például aspermából, sőt a székletből is.<ref name="NEJM-FirstUS" /><ref name="Semen" /> |
|||
==Kutatási területei== |
|||
Munkatársaival kidolgozta több enzim, így a pepszin, a dezoxiribonukleáz és más enzimek általánosan alkalmazható kristályosításának eljárását. Egyértelműen bizonyította, hogy ezek fehérjék. Módszereinek felhasználásával kristályosította [[Wendell M. Stanley|Wendell Stanley]] a dohánymozaikvírust. |
|||
A kórokozó fertőzőképessége a betegség, illetve a korai, tünetmentes szakasz alatt még nem teljesen ismert, de a jelenlegi adatok szerint a torokban a virionszám nagyjából a fertőzés utáni negyedik napon<ref name="NaturePeakLoad" /><ref name="ScienceFlawed" /> vagy tünetek megjelenése utáni első héten a legmagasabb, utána pedig fokozatosan csökken.<ref name="LancetLoad" /> A WHO első megállapításaival ellentétben<ref name="WHO-SR12" /> az epidemiológiai modellek arra utalnak, hogy a teljesen tünetmentes, illetve korai fázisban lévő betegek az új fertőzések legfőbb forrásai.<ref name="ScienceRapid" /> Egy Montevideóban kikötött óceánjáró 217 utasa és legénysége közül 128-nak lett pozitív a tesztje, míg tüneteket csak 24-en észleltek.<ref name="Thorax" /> Egy 94 betegen elvégzett vizsgálat arra utal, hogy leginkább 2-3 nappal a tünetek megjelenése előtt fertőzőképesek.<ref name="NatureTempShedding" /> |
|||
==Szakmai sikerek== |
|||
* [[1946]]-ban James Sumner és [[Wendell Stanley]] társaságában megosztott [[kémiai Nobel-díj]]at kaptak az [[enzim]]ek és [[vírus]]fehérjék tiszta formában történő előállításáért. |
|||
Ritka esetekben előfordul, hogy a vírus emberről állatra terjed át, például macskákra,<ref name="oie.int" /><ref name="BronxTiger" /> emiatt egyes intézmények azt javasolják, hogy a betegek lehetőleg ne érintkezzenek háziállatokkal.<ref name="USDAtiger" /><ref name="CDCanimals" /> |
|||
== Jegyzetek == |
|||
{{jegyzetek}} |
|||
== |
==Eredete== |
||
[[File:SARS-CoV-1 and 2 - mammals as carriers.png|thumb|upright=1.25|Transmission of SARS-CoV-1 and SARS-CoV-2 from mammals as biological carriers to humans]] |
|||
*{{cite web|url=http://www.kfki.hu/physics/historia/historia/egyen.php?namenev=northrop&nev5=Northrop,+John+H. |
|||
A SARS-CoV-2-fertőzés első eseteit a kínai Vuhan városában észlelték.<ref name="NatureZhou" /> Az állatról emberre való átadódás körülményei egyelőre tisztázatlanok.<ref name="early" /><ref name="PopSciJan" /><ref name="Proximal" /> A betegek közül sokan a vuhani Huanan élelmiszerpiac dolgozói voltak,<ref name="LancetClinical" /><ref name="LancetCharacteristics" /> ezért feltételezik, hogy a humán patogén törzs itt alakulhatott ki.<ref name="Proximal" /><ref name="nature feb2020" /> Nem zárható ki azonban, hogy a piacra is kívülről került a vírus és csak itt kezdett gyors terjedésbe.<ref name="early" /><ref name="XivDecoding" /> A korai megbetegedésekből (2019 december-2020 február) származó 160 minta alapján a SARS-CoV-2 olyan denevér-koronavírusokhoz hasonlít a leginkább, amelyek Kanton tartományban a leggyakoribbak.<ref name="PNAS2020" /><ref name="CU2020" /> |
|||
|title=John Howard Northrop|accessdate=2012-5-27|publisher=kfki.hu}} |
|||
{{KémiaiNobelDíj}} |
|||
{{nemzetközi katalógusok}} |
|||
{{portál|kémia||USA|-}} |
|||
{{DEFAULTSORT:Northrop, John Howard}} |
|||
A 2002-2004-es SARS-járvány után átfogó kutatás indult a hasonló, állatokban élő vírusok után és kimutatták, hogy számos denevérfaj, elsősorban a parkósorrú denevérek (a ''Rhinolophus'' nemzetség tagjai) hordoznak hasonló koronavírusokat. A ''Rhinolophus sinicus'' egyik vírusa 80%-os,<ref name="MedVirEvolution" /><ref name="NCBI-Bat" /><ref name="NCBI-Bat2" />míg a ''Rhinolophus affinis'' vírustörzse 96%-os hasonlóságot mutatott a SARS-CoV-2-vel.<ref name="NatureZhou" /><ref name="NCBI-Bat3" /> |
|||
[[xKategória:1891-ben született személyek]] |
|||
[[xKategória:1987-ben elhunyt személyek]] |
|||
[[File:Naturalis Biodiversity Center - RMNH.MAM.33160.b dor - Rhinolophus sinicus - skin.jpeg|thumb|left|Samples taken from ''Rhinolophus sinicus'', a species of [[horseshoe bat]]s, show a 80% resemblance to SARS-CoV-2.]] |
|||
[[xKategória:Amerikai kémikusok]] |
|||
Bats were initially considered to be the most likely natural reservoir of SARS-CoV-2,<ref name="WHOChinaJoint" /><ref name="LancetBinding" /> which means that they harbour the virus for long periods of time with no pathogenic effects. Regarding the animal source of infection into humans, the differences between the bat coronavirus sampled at the time and SARS-CoV-2 suggested that humans were infected via an intermediate host. Arinjay Banerjee, a virologist at [[McMaster University]], notes that "the [[Severe acute respiratory syndrome coronavirus|SARS virus]] shared 99.8% of its [[genome]] with a [[civet]] coronavirus, which is why civets were considered the source."<ref name="nature feb2020" /> Although studies had suggested some likely candidates, the number and identities of intermediate hosts remains uncertain.<ref name="IJID-interm-host" /> Nearly half of the strain's genome had a phylogenetic lineage distinct from known relatives.<ref name="Rejects" /> |
|||
[[xKategória:Amerikai biokémikusok]] |
|||
[[xKategória:Nobel-díjas kémikusok]] |
|||
[[File:Zoo Leipzig - Tou Feng.jpg|thumb|right|alt=Chinese pangolin|The [[pangolin]] coronavirus has up to 92% resemblance to SARS-CoV-2.<ref name="CurrentOrigin" />]] |
|||
[[xKategória:Amerikai Nobel-díjasok]] |
|||
A [[phylogenetics]] study published in 2020 indicates that [[pangolin]]s are a reservoir host of SARS-CoV-2-like coronaviruses.<ref name="BMJ-Best-Practice" /> However, there is no direct evidence to link pangolins as an intermediate host of SARS-CoV-2 at this moment. While there is scientific consensus that bats are the ultimate source of coronaviruses, it is hypothesized that a SARS-CoV-2-like coronavirus originated in pangolins, jumped back to bats, and then jumped to humans, resulting in SARS-CoV-2. Based on whole genome sequence similarity, a pangolin coronavirus candidate strain was found to be less similar than RaTG13, but more similar than other bat coronaviruses to SARS-CoV-2.<ref name="CurrentOrigin" /> Therefore, based on [[maximum parsimony]], a specific population of bats is more likely to have directly transmitted SARS-CoV-2 to humans than a pangolin, while an evolutionary ancestor to bats was the source of general coronaviruses.<ref name="ForbesOrigin" /> |
|||
[[xKategória:Amerikai egyetemi, főiskolai oktatók]] |
|||
A [[metagenomics]] study published in 2019 had previously revealed that SARS-CoV, the strain of the virus that causes SARS, was the most widely distributed coronavirus among a sample of [[Sunda pangolin]]s.<ref name="VirusesPangolins" /> On 7{{nbsp}}February 2020, [[South China Agricultural University]] in [[Guangzhou]] announced that researchers discovered a pangolin sample with a particular coronavirus – a single [[nucleic acid]] sequence of the virus was "99% similar" to that of a [[protein]]-coding [[RNA]] of SARS-CoV-2.<ref name="NaturePang" /> The authors state that "the receptor-binding domain of the [[Peplomer|S protein]] [that binds to the [[cell surface receptor]] during infection] of the newly discovered Pangolin-CoV is virtually identical to that of 2019-nCoV, with one [[amino acid]] difference."<ref name="Isolation" /> Microbiologists and geneticists in [[Texas]] have independently found evidence of [[reassortment]] in coronaviruses suggesting involvement of pangolins in the origin of SARS-CoV-2.<ref name="WongRecombination" /> The majority of the viral RNA is related to a variation of bat coronaviruses. The spike protein appears to be a notable exception, however, possibly acquired through a more recent recombination event with a pangolin coronavirus.<ref>{{Cite web|last=Timmer|first=John|date=1 June 2020|title=SARS-CoV-2 looks like a hybrid of viruses from two different species|url=https://arstechnica.com/science/2020/06/sars-cov-2-looks-like-a-hybrid-of-viruses-from-two-different-species/|access-date=6 June 2020|website=Ars Technica|language=en-us|archive-url=https://web.archive.org/web/20200605181132/https://arstechnica.com/science/2020/06/sars-cov-2-looks-like-a-hybrid-of-viruses-from-two-different-species/|archive-date=5 June 2020|url-status=live}}</ref> Structural analysis of the receptor binding domain (RBD) and human [[angiotensin-converting enzyme 2]] (ACE2) complex<ref>{{Cite journal|last1=Yan|first1=Renhong|last2=Zhang|first2=Yuanyuan|last3=Li|first3=Yaning|last4=Xia|first4=Lu|last5=Guo|first5=Yingying|last6=Zhou|first6=Qiang|date= 27 March 2020|title=Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2|journal=Science|volume=367|issue=6485|pages=1444–1448|doi=10.1126/science.abb2762|issn=1095-9203|pmc=7164635|pmid=32132184|bibcode=2020Sci...367.1444Y}}</ref> revealed key mutations on the RBD, such as F486 and N501, which form contacts with ACE2.<ref name=":0">{{Cite journal|last=Ho|first=Mitchell|s2cid=219476100|date=2020-04-30|title=Perspectives on the development of neutralizing antibodies against SARS-CoV-2|url=https://academic.oup.com/abt/article/3/2/109/5841095|journal=Antibody Therapeutics|language=en|volume=3|issue=2|pages=109–114|doi=10.1093/abt/tbaa009|pmid=32566896|pmc=7291920|access-date=14 June 2020|archive-url=https://web.archive.org/web/20200614155711/https://academic.oup.com/abt/article/3/2/109/5841095|archive-date=14 June 2020|url-status=live}}</ref> These residues are found in the pangolin coronavirus.<ref name=":0" /> |
|||
Pangolins are protected under Chinese law, but their [[pangolin trade|poaching and trading]] for use in [[traditional Chinese medicine]] remains common in the [[black market]].<ref name="TelegraphPangolins" /><ref name="NYT-Ban" /> [[Deforestation]], wildlife farming and trade in unsanitary conditions increases the risk of new zoonotic diseases, biodiversity experts have warned.<ref>{{Cite news|last=Carrington|first=Damian|date=27 April 2020|title=Halt destruction of nature or suffer even worse pandemics, say world's top scientists|language=en-GB|work=The Guardian|url=https://www.theguardian.com/world/2020/apr/27/halt-destruction-nature-worse-pandemics-top-scientists|url-status=live|access-date=31 May 2020|issn=0261-3077|archive-url=https://web.archive.org/web/20200515015940/https://www.theguardian.com/world/2020/apr/27/halt-destruction-nature-worse-pandemics-top-scientists|archive-date=15 May 2020}}</ref><ref>{{Cite web|title=How deforestation can lead to more infectious diseases|url=https://www.dw.com/en/how-deforestation-can-lead-to-more-infectious-diseases/a-53282244|last=Pontes|first=Nadia|date=29 April 2020|website=DW.COM|language=en-GB|url-status=live|archive-url=https://web.archive.org/web/20200505160903/https://www.dw.com/en/how-deforestation-can-lead-to-more-infectious-diseases/a-53282244|archive-date=5 May 2020|access-date=31 May 2020}}</ref><ref>{{Cite journal|last1=Cheng|first1=Vincent C. C.|last2=Lau|first2=Susanna K. P.|last3=Woo|first3=Patrick C. Y.|last4=Yuen|first4=Kwok Yung|date=October 2007|title=Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection|journal=Clinical Microbiology Reviews|volume=20|issue=4|pages=660–694|doi=10.1128/CMR.00023-07|issn=0893-8512|pmc=2176051|pmid=17934078}}</ref> |
|||
It is unlikely that SARS-CoV-2 was [[Genetic engineering|genetically engineered]]. According to [[Computer simulation|computational simulations]] on [[protein folding]], the RBD of the spike protein of SARS-CoV-2 should have unremarkable binding affinity. In actuality, however, it has very efficient binding to the human ACE2 receptor. To expose the RBD for fusion, [[furin]] [[protease]]s must first cleave the S protein. Furin proteases are abundant in the respiratory tract and lung epithelial cells. Additionally, the backbone of the virus does not resemble any previously described in scientific literature used for genetic modification. The possibility that the virus could have gained the necessary [[adaptation]]s through [[cell culture]] in a laboratory setting is challenged by scientists who assert that "the generation of the predicted [[O-linked glycosylation|O-linked glycans]]... suggest[s] the involvement of an [[immune system]]."<ref name="EA-20200317" /><ref name="Proximal" /> |
|||
===Phylogenetics and taxonomy=== |
|||
{{Infobox genome |
|||
| image = File:SARS-CoV-2 genome.svg |
|||
| caption = [[Genomic]] organisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2 |
|||
| taxId = 86693 |
|||
| size = 29,903 bases |
|||
| year = 2020 |
|||
| ucsc_assembly = wuhCor1 |
|||
}} |
|||
SARS-CoV-2 belongs to the broad family of viruses known as [[coronavirus]]es.<ref name="Fox2020" /> It is a [[Positive-sense single-stranded RNA virus|positive-sense single-stranded RNA]] (+ssRNA) virus, with a single linear RNA segment. Other coronaviruses are capable of causing illnesses ranging from the [[common cold]] to more severe diseases such as [[Middle East respiratory syndrome]] (MERS, fatality rate ~34%). It is the seventh known coronavirus to infect people, after [[Human coronavirus 229E|229E]], [[Human coronavirus NL63|NL63]], [[Human coronavirus OC43|OC43]], [[Human coronavirus HKU1|HKU1]], [[Middle East respiratory syndrome-related coronavirus|MERS-CoV]], and the original [[Severe acute respiratory syndrome coronavirus|SARS-CoV]].<ref name="NEJM-Novel" /> |
|||
Like the SARS-related coronavirus strain implicated in the 2003 SARS outbreak, SARS-CoV-2 is a member of the subgenus ''[[Sarbecovirus]]'' ([[beta-CoV]] lineage B).<ref name="NextstrainPhylogeny" /><ref name="Wong2019" /> Its RNA sequence is approximately 30,000 [[nucleobase|base]]s in length.<ref name="gisaid" /> SARS-CoV-2 is unique among known betacoronaviruses in its incorporation of a [[polybasic cleavage site]], a characteristic known to increase [[pathogenicity]] and transmissibility in other viruses.<ref name="Proximal" /><ref name="CellWalls" /><ref name="AntiviralCleavage" /> |
|||
With a sufficient number of sequenced [[genome]]s, it is possible to reconstruct a [[phylogenetic tree]] of the mutation history of a family of viruses. By 12 January 2020, five genomes of SARS-CoV-2 had been isolated from Wuhan and reported by the [[Chinese Center for Disease Control and Prevention]] (CCDC) and other institutions;<ref name="gisaid" /><ref name="VirologicalInitial" /> the number of genomes increased to 42 by 30 January 2020.<ref name="NextstrainJanuary" /> A phylogenetic analysis of those samples showed they were "highly related with at most seven mutations relative to a [[common ancestor]]", implying that the first human infection occurred in November or December 2019.<ref name="NextstrainJanuary" /> {{As of|2020|May|7|post=,}} 4,690 SARS-CoV-2 genomes sampled on six continents were publicly available.<ref name="NexstrainApril" /> |
|||
On 11 February 2020, the International Committee on Taxonomy of Viruses announced that according to existing rules that compute hierarchical relationships among coronaviruses on the basis of five [[conserved sequence]]s of nucleic acids, the differences between what was then called 2019-nCoV and the virus strain from the 2003 SARS outbreak were insufficient to make them separate [[viral species]]. Therefore, they identified 2019-nCoV as a [[Strain (biology)|strain]] of ''[[Severe acute respiratory syndrome-related coronavirus]]''.<ref name="CoronavirusStudyGroup" /> |
|||
===Structural biology=== |
|||
[[File:Coronavirus virion structure.svg|alt=Figure of a spherical SARSr-CoV virion showing locations of structural proteins forming the viral envelope and the inner nucleocapsid|thumb|right|Structure of a [[SARSr-CoV]] virion]] |
|||
Each SARS-CoV-2 [[virion]] is 50–200 [[nanometre]]s in diameter.<ref name="LancetCharacteristics" /> Like other coronaviruses, SARS-CoV-2 has four structural proteins, known as the S ([[Peplomer|spike]]), E (envelope), M ([[membrane]]), and N ([[nucleocapsid]]) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the [[viral envelope]].<ref name="WuStructure" /> The spike protein, which has been imaged at the atomic level using [[cryogenic electron microscopy]],<ref name="SCI-20200219" /><ref name="GZM-20200220" /> is the protein responsible for allowing the virus to attach to and fuse with the [[cell membrane|membrane]] of a host cell;<ref name="WuStructure" /> specifically, its S1 subunit catalyzes attachment, the S2 subunit fusion.<ref name="CEBMcoronaviruses" /> |
|||
[[File:6VSB spike protein SARS-CoV-2 monomer in homotrimer.png|thumb|upright|alt=SARS-CoV-2 spike homotrimer focusing upon one protein subunit with an ACE2 binding domain highlighted|SARS-CoV-2 spike [[homotrimer]] with one [[protein subunit]] highlighted. The ACE2 [[binding domain]] is magenta.]] |
|||
[[Protein structure prediction|Protein modeling]] experiments on the spike protein of the virus soon suggested that SARS-CoV-2 has sufficient affinity to the receptor [[angiotensin converting enzyme 2]] (ACE2) on human cells to use them as a mechanism of [[Viral entry|cell entry]].<ref name="SCLSModeling" /> By 22 January 2020, a group in China working with the full virus genome and a group in the United States using [[reverse genetics]] methods independently and experimentally demonstrated that ACE2 could act as the receptor for SARS-CoV-2.<ref name="NatureZhou" /><ref name="Letko22Jan2020" /><ref name="NatMicLetko" /><ref name="ElSahly" /> Studies have shown that SARS-CoV-2 has a higher affinity to human ACE2 than the original SARS virus strain.<ref name="SCI-20200219" /><ref name="NIH-Structure" /> SARS-CoV-2 may also use [[basigin]] to assist in cell entry.<ref name="CD147" /> |
|||
Initial spike protein priming by [[TMPRSS2|transmembrane protease, serine 2]] (TMPRSS2) is essential for entry of SARS-CoV-2.<ref name="HoffmanCell" /> After a SARS-CoV-2 virion attaches to a target cell, the cell's [[protease]] TMPRSS2 cuts open the spike protein of the virus, exposing a [[fusion peptide]] in the S2 subunit, and the host receptor ACE2.<ref name="CEBMcoronaviruses" /> After fusion, an [[endosome]] forms around the virion, separating it from the rest of the host cell. The virion escapes when the [[pH]] of the endosome drops or when [[cathepsin]], a host [[cysteine]] protease, cleaves it.<ref name="CEBMcoronaviruses" /> The virion then releases RNA into the cell and forces the cell to produce and disseminate [[Viral replication|copies of the virus]], which infect more cells.<ref name="econ-anatomy-killer" /> |
|||
SARS-CoV-2 produces at least three [[virulence factor]]s that promote shedding of new virions from host cells and inhibit [[immune response]].<ref name="WuStructure" /> Whether they include [[downregulation]] of ACE2, as seen in similar coronaviruses, remains under investigation (as of May 2020).<ref name="BMJ-Best-Practice" /> |
|||
{{Multiple image | align = center | direction = horizontal | total_width = 500 | image1 = SARS-CoV-2 49531042877.jpg | alt1 = SARS-CoV-2 emerging from a human cell | image2 = SARS-CoV-2 scanning electron microscope image.jpg | alt2 = SARS-CoV-2 virions emerging from a human cell | footer_align = center | footer = Digitally colourised [[scanning electron micrographs]] of SARS-CoV-2 [[virion]]s (yellow) emerging from human cells [[Cell culture|cultured]] in a laboratory}} |
|||
==Epidemiology== |
|||
{{Main|COVID-19 pandemic}} |
|||
[[File:Novel Coronavirus SARS-CoV-2 (49597020718).jpg|thumb|[[Transmission electron micrograph]] of SARS-CoV-2 virions (red) isolated from a patient during the [[COVID-19 pandemic]]|alt=Micrograph of SARS-CoV-2 virus particles isolated from a patient]] |
|||
Based on the low variability exhibited among known SARS-CoV-2 [[genomic]] sequences, the strain is thought to have been detected by health authorities within weeks of its emergence among the human population in late 2019.<ref name="early" /><ref name="FromHere" /> The earliest case of infection currently known is dated back to 17 November 2019 or possibly 1 December 2019.<ref name="November case" /> The virus subsequently spread to all provinces of China and to more than 150 other countries in Asia, Europe, North America, South America, Africa, and Oceania.<ref name="JHU_ticker" /> Human-to-human transmission of the virus has been confirmed in all these regions.<ref name="WHO-SR69" /> On 30 January 2020, SARS-CoV-2 was designated a [[Public Health Emergency of International Concern]] by the WHO,<ref name="NYT 20200130" /><ref name="WHO-PHEIC" /> and on 11 March 2020 the WHO declared it a [[pandemic]].<ref name="WHOPandemic" /><ref name="WSJPandemic" /> |
|||
The [[basic reproduction number]] (<math>R_0</math>) of the virus has been estimated to be between 1.4 and 3.9.<ref name="NEJM-Dynamics" /><ref name="Eurosurveillance" /> This means each infection from the virus is expected to result in 1.4 to 3.9 new infections when no members of the community are [[immunity (medical)|immune]] and no [[infection control|preventive measures]] are taken. The reproduction number may be higher in densely populated conditions such as those found on [[cruise ship]]s.<ref name="Rocklov" /> Many forms of [[coronavirus disease 2019#Prevention|preventive efforts]] may be employed in specific circumstances in order to reduce the propagation of the virus. |
|||
There have been about 82,000 confirmed cases of infection in mainland China.<ref name="JHU_ticker" /> While the proportion of infections that result in [[COVID-19 pandemic#Cases|confirmed cases]] or progress to diagnosable disease remains unclear,<ref name="STAT-Severity" /> one mathematical model estimated that 75,815 people were infected on 25 January 2020 in Wuhan alone, at a time when the number of confirmed cases worldwide was only 2,015.<ref name="pmid32014114" /> Before 24 February 2020, over 95% of all deaths from [[COVID-19]] worldwide had occurred in [[Hubei|Hubei province]], where Wuhan is located.<ref name="GuardianLeap" /><ref name="SunChinaAfrica" /> As of {{Cases in the COVID-19 pandemic|date|editlink=|ref=no}}, the percentage had decreased to {{percentage|3216|{{Cases in the COVID-19 pandemic|deaths|editlink=no|ref=no}}|sigfig=2}}.{{Cases in the COVID-19 pandemic|ref=yes}} |
|||
As of {{Cases in the COVID-19 pandemic|date|editlink=|ref=no}}, there have been {{Cases in the COVID-19 pandemic|confirmed|editlink=no|ref=no}} total confirmed cases of SARS-CoV-2 infection in the ongoing pandemic.{{Cases in the COVID-19 pandemic|ref=yes}} The total number of deaths attributed to the virus is {{Cases in the COVID-19 pandemic|deaths|editlink=no|ref=no}}.{{Cases in the COVID-19 pandemic|ref=yes}} Many recoveries from confirmed infections go unreported, but at least {{Cases in the COVID-19 pandemic|recovered|editlink=no|ref=no}} people have recovered from confirmed infections.{{Cases in the COVID-19 pandemic|ref=yes}} |
|||
{{clear}} |
|||
==See also== |
|||
* ''[[Decoding COVID-19]]'' – 2020 PBS film documentary about the 2019–2020 COVID-19 pandemic |
|||
{{Portal bar|border=no|Coronavirus disease 2019|Medicine|Viruses}} |
|||
==References== |
|||
<!--This article uses [[Help:List-defined references]] (LDR). Please place the full links below in alphabetical order.--> |
|||
{{reflist|refs= |
|||
<ref name="AntiviralCleavage">{{Cite journal |vauthors=Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E |date=February 2020 |title=The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade |journal=[[Antiviral Research]] |volume=176 |page=104742 |doi=10.1016/j.antiviral.2020.104742 |pmc=7114094 |pmid=32057769 |name-list-format=vanc}}</ref> |
|||
<ref name="AtlanticSuccess">{{Cite web |url=https://www.theatlantic.com/science/archive/2020/03/biography-new-coronavirus/608338/ |title=Why the Coronavirus Has Been So Successful |date=20 March 2020 |website=The Atlantic |url-status=live |archive-url=https://web.archive.org/web/20200320161836/https://www.theatlantic.com/science/archive/2020/03/biography-new-coronavirus/608338/ |archive-date=20 March 2020 |access-date=20 March 2020 |vauthors=Yong E |name-list-format=vanc}}</ref> |
|||
<ref name="BBC-Named">{{Cite news |url=https://www.bbc.com/news/world-asia-china-51466362 |title=Coronavirus disease named Covid-19 |date=11 February 2020 |work=[[BBC News Online]] |access-date=15 February 2020 |url-status=live |archive-url=https://web.archive.org/web/20200215204154/https://www.bbc.com/news/world-asia-china-51466362 |archive-date=15 February 2020 |name-list-format=vanc}}</ref> |
|||
<!-- <ref name="BloombergMarch2020">{{Cite web|title=Trump Says He'll Stop Using the Term 'Chinese Virus'|url=https://www.bloomberg.com/news/articles/2020-03-25/trump-says-he-ll-stop-using-chinese-virus-easing-blame-game|last=Leigh|first=Karen|date=24 March 2020|website=Bloomberg|url-status=live|archive-url=https://web.archive.org/web/20200401223041/https://www.bloomberg.com/news/articles/2020-03-25/trump-says-he-ll-stop-using-chinese-virus-easing-blame-game|archive-date=1 April 2020|access-date=7 May 2020}}</ref>--> |
|||
<ref name="BMJ-Best-Practice">{{cite web |first1=Nicholas J |last1=Beeching |first2=Tom E |last2=Fletcher |first3=Robert |last3=Fowler |url=https://bestpractice.bmj.com/topics/en-us/3000168/pdf/3000168.pdf |access-date=25 May 2020 |title=BMJ Best Practice: Coronavirus Disease 2019 (COVID-19) |website=[[BMJ]] |language=en |date=22 May 2020 |name-list-format=vanc |archive-url=https://web.archive.org/web/20200613123054/https://bestpractice.bmj.com/topics/en-us/3000168/pdf/3000168.pdf |archive-date=13 June 2020 |url-status=live }}</ref> |
|||
<ref name="BronxTiger">{{Cite news |last=Goldstein |first=Joseph |url=https://www.nytimes.com/2020/04/06/nyregion/bronx-zoo-tiger-coronavirus.html |title=Bronx Zoo Tiger Is Sick with the Coronavirus |date=6 April 2020 |work=[[The New York Times]] |access-date=10 April 2020 |name-list-format=vanc |archive-url=https://web.archive.org/web/20200409094808/https://www.nytimes.com/2020/04/06/nyregion/bronx-zoo-tiger-coronavirus.html |archive-date=9 April 2020 |url-status=live}}</ref> |
|||
<ref name="CD147">{{Cite journal |displayauthors=6 |vauthors=Wang K, Chen W, Zhou YS, Lian JQ, Zhang Z, Du P, Gong L, Zhang Y, Cui HY, Geng JJ, Wang B, Sun XX, Wang CF, Yang X, Lin P, Deng YQ, Wei D, Yang XM, Zhu YM, Zhang K, Zheng ZH, Miao JL, Guo T, Shi Y, Zhang J, Fu L, Wang QY, Bian H, Zhu P, Chen ZN |s2cid=214725955 |date=14 March 2020 |title=SARS-CoV-2 invades host cells via a novel route: CD147-spike protein |journal=[[bioRxiv]] |type=preprint |doi=10.1101/2020.03.14.988345 |name-list-format=vanc |url=https://www.biorxiv.org/content/biorxiv/early/2020/03/14/2020.03.14.988345.full.pdf |access-date=5 May 2020 |archive-url=https://web.archive.org/web/20200511222955/https://www.biorxiv.org/content/biorxiv/early/2020/03/14/2020.03.14.988345.full.pdf |archive-date=11 May 2020 |url-status=live}}</ref> |
|||
<ref name="CDC-nCoV">{{Cite web |url=https://www.cdc.gov/coronavirus/2019-ncov/hcp/faq.html |title=Healthcare Professionals: Frequently Asked Questions and Answers |date=11 February 2020 |website=United States [[Centers for Disease Control and Prevention]] (CDC) |url-status=live |archive-url=https://web.archive.org/web/20200214023335/https://www.cdc.gov/coronavirus/2019-ncov/hcp/faq.html |archive-date=14 February 2020 |access-date=15 February 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="CDCAbout">{{Cite web |url=https://www.cdc.gov/coronavirus/2019-ncov/about/index.html |title=About Novel Coronavirus (2019-nCoV) |date=11 February 2020 |website=United States [[Centers for Disease Control and Prevention]] (CDC) |url-status=live |archive-url=https://web.archive.org/web/20200211105920/https://www.cdc.gov/coronavirus/2019-ncov/about/index.html |archive-date=11 February 2020 |access-date=25 February 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="CDCanimals">{{Cite web |url=https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/animals.html |title=If You Have Animals—Coronavirus Disease 2019 (COVID-19) |date=13 April 2020 |website=[[Centers for Disease Control and Prevention]] (CDC) |language=en-us |access-date=16 April 2020 |archive-url=https://web.archive.org/web/20200401163720/https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/animals.html |archive-date=1 April 2020 |url-status=live}}</ref> |
|||
<ref name="CDCTrans">{{Cite web |url=https://www.cdc.gov/coronavirus/2019-ncov/about/transmission.html |title=How COVID-19 Spreads |date=27 January 2020 |website=U.S. [[Centers for Disease Control and Prevention]] (CDC) |url-status=live |archive-url=https://web.archive.org/web/20200128152653/https://www.cdc.gov/coronavirus/2019-ncov/about/transmission.html |archive-date=28 January 2020 |access-date=29 January 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="CEBMcoronaviruses">{{Cite web |last1=Aronson |first1=Jeffrey K |title=Coronaviruses – a general introduction |date=25 March 2020 |access-date=24 May 2020 |url=https://www.cebm.net/covid-19/coronaviruses-a-general-introduction/ |website=Centre for Evidence-Based Medicine, Nuffield Department of Primary Care Health Sciences, University of Oxford |name-list-format=vanc |archive-url=https://web.archive.org/web/20200522053938/https://www.cebm.net/covid-19/coronaviruses-a-general-introduction/ |archive-date=22 May 2020 |url-status=live }}</ref> |
|||
<ref name="CellWalls">{{Cite journal |vauthors=Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D |date=9 March 2020 |title=Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein |journal=[[Cell (journal)|Cell]] |volume=181 |issue=2 |pages=281–292.e6 |doi=10.1016/j.cell.2020.02.058 |pmc=7102599 |pmid=32155444 |name-list-format=vanc}}</ref> |
|||
<ref name="Chan24Jan2020">{{Cite journal |displayauthors=6 |vauthors=Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY |date=February 2020 |title=A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster |journal=[[The Lancet]] |volume=395 |issue=10223 |pages=514–523 |doi=10.1016/S0140-6736(20)30154-9 |pmc=7159286 |pmid=31986261 |name-list-format=vanc}}</ref> |
|||
<ref name="CoronavirusStudyGroup">{{Cite journal |displayauthors=6 |vauthors=Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, ((Poon LLM)), Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J |date=March 2020 |title=The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2 |journal=[[Nature Microbiology]] |volume=5 |issue=4 |pages=536–544 |doi=10.1038/s41564-020-0695-z |pmc=7095448 |pmid=32123347 |name-list-format=vanc}}</ref> |
|||
<ref name="COVIDname">{{Cite web |url=https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it |title=Naming the coronavirus disease (COVID-2019) and the virus that causes it |publisher=World Health Organization |url-status=live |archive-url=https://web.archive.org/web/20200228035651/https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it |archive-date=28 February 2020 |access-date=24 February 2020 |quote=From a risk communications perspective, using the name SARS can have unintended consequences in terms of creating unnecessary fear for some populations.{{nbsp}}... For that reason and others, WHO has begun referring to the virus as "the virus responsible for COVID-19" or "the COVID-19 virus" when communicating with the public. Neither of these designations are [sic] intended as replacements for the official name of the virus as agreed by the ICTV. |name-list-format=vanc}}</ref> |
|||
<ref name="CU2020">{{Cite web |url=https://www.cam.ac.uk/research/news/covid-19-genetic-network-analysis-provides-snapshot-of-pandemic-origins |title=COVID-19: genetic network analysis provides 'snapshot' of pandemic origins |date=9 April 2020 |website=Cambridge University |language=en |access-date=17 April 2020 |archive-url=https://web.archive.org/web/20200416210214/https://www.cam.ac.uk/research/news/covid-19-genetic-network-analysis-provides-snapshot-of-pandemic-origins |archive-date=16 April 2020 |url-status=live}}</ref> |
|||
<ref name="CurrentOrigin">{{Cite journal |vauthors=Zhang T, Wu Q, Zhang Z |date=19 March 2020 |title=Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak |journal=[[Current Biology]] |volume=30 |issue=7 |pages=1346–1351.e2 |doi=10.1016/j.cub.2020.03.022 |pmc=7156161 |pmid=32197085}}</ref> |
|||
<ref name="DWPangolins">{{Cite news |url=https://www.dw.com/en/coronavirus-from-bats-to-pangolins-how-do-viruses-reach-us/a-52291570 |title=Coronavirus: From bats to pangolins, how do viruses reach us? |date=7 February 2020 |access-date=13 March 2020 |publisher=[[Deutsche Welle]] |vauthors=Shield C |name-list-format=vanc |archive-url=https://web.archive.org/web/20200604011742/https://www.dw.com/en/coronavirus-from-bats-to-pangolins-how-do-viruses-reach-us/a-52291570 |archive-date=4 June 2020 |url-status=live }}</ref> |
|||
<ref name="EA-20200317">{{cite news |work=[[EurekAlert!]] |publisher=Scripps Research Institute |title=The COVID-19 coronavirus epidemic has a natural origin, scientists say—Scripps Research's analysis of public genome sequence data from SARS‑CoV‑2 and related viruses found no evidence that the virus was made in a laboratory or otherwise engineered |url=https://www.eurekalert.org/pub_releases/2020-03/sri-tcc031720.php |date=17 March 2020 |accessdate=15 April 2020 |author-link=Scripps Research Institute |archive-url=https://web.archive.org/web/20200403083606/https://www.eurekalert.org/pub_releases/2020-03/sri-tcc031720.php |archive-date=3 April 2020 |url-status=live}}</ref> |
|||
<ref name="early">{{Cite journal |vauthors=Cohen J |date=January 2020 |title=Wuhan seafood market may not be source of novel virus spreading globally |journal=[[Science (journal)|Science]] |doi=10.1126/science.abb0611 |name-list-format=vanc}}</ref> |
|||
<ref name="econ-anatomy-killer">{{Cite magazine |date=12 March 2020 |title=Anatomy of a Killer: Understanding SARS-CoV-2 and the drugs that might lessen its power |url=https://www.economist.com/briefing/2020/03/12/understanding-sars-cov-2-and-the-drugs-that-might-lessen-its-power |magazine=[[The Economist]] |archive-url=https://web.archive.org/web/20200314010231/https://www.economist.com/briefing/2020/03/12/understanding-sars-cov-2-and-the-drugs-that-might-lessen-its-power |archive-date=14 March 2020 |access-date=14 March 2020 |url-status=live |name-list-format=vanc}}</ref> |
|||
<ref name="EconomistSinophobia">{{cite news|author= |title= The coronavirus spreads racism against—and among—ethnic Chinese|url= https://www.economist.com/china/2020/02/17/the-coronavirus-spreads-racism-against-and-among-ethnic-chinese|work= [[The Economist]]|date= 17 February 2020|access-date= 17 February 2020|archive-url= https://web.archive.org/web/20200217223902/https://www.economist.com/china/2020/02/17/the-coronavirus-spreads-racism-against-and-among-ethnic-chinese|archive-date= 17 February 2020|url-status= live| name-list-format = vanc}}</ref> |
|||
<ref name="ElSahly">{{Cite journal |vauthors=((El Sahly HM)) |title=Genomic Characterization of the 2019 Novel Coronavirus |url=https://www.jwatch.org/na50823/2020/02/06/genomic-characterization-2019-novel-coronavirus |url-status=live |journal=[[The New England Journal of Medicine]] |archive-url=https://web.archive.org/web/20200217184052/https://www.jwatch.org/na50823/2020/02/06/genomic-characterization-2019-novel-coronavirus |archive-date=17 February 2020 |access-date=9 February 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="Eurosurveillance">{{Cite journal |vauthors=Riou J, Althaus CL |date=January 2020 |title=Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020 |journal=[[Eurosurveillance]] |volume=25 |issue=4 |doi=10.2807/1560-7917.ES.2020.25.4.2000058 |pmc=7001239 |pmid=32019669 |name-list-format=vanc}}</ref> |
|||
<ref name="ForbesOrigin">{{Cite web |url=https://www.forbes.com/sites/coronavirusfrontlines/2020/04/17/a-virologist-explains-why-it-is-unlikely-covid-19-escaped-from-a-lab/ |title=A Virologist Explains Why It Is Unlikely COVID-19 Escaped From A Lab |vauthors=Kindrachuk J, ((Coronavirus Frontlines)) |work=Forbes |access-date=22 April 2020 |date=17 April 2020 |name-list-format=vanc |archive-url=https://web.archive.org/web/20200421093833/https://www.forbes.com/sites/coronavirusfrontlines/2020/04/17/a-virologist-explains-why-it-is-unlikely-covid-19-escaped-from-a-lab/ |archive-date=21 April 2020 |url-status=live}}</ref> |
|||
<ref name="Fox2020">{{Cite journal |vauthors=Fox D |date=24 January 2020 |title=What you need to know about the Wuhan coronavirus |journal=[[Nature (journal)|Nature]] |doi=10.1038/d41586-020-00209-y |name-list-format=vanc}}</ref> |
|||
<ref name="FromHere">{{Cite web |url=https://www.genengnews.com/insights/what-we-know-today-about-coronavirus-sars-cov-2-and-where-do-we-go-from-here/ |title=What We Know Today about Coronavirus SARS-CoV-2 and Where Do We Go from Here |date=19 February 2020 |website=[[Mary Ann Liebert|Genetic Engineering and Biotechnology News]] |url-status=live |archive-url=https://web.archive.org/web/20200314074654/https://www.genengnews.com/insights/what-we-know-today-about-coronavirus-sars-cov-2-and-where-do-we-go-from-here/ |archive-date=14 March 2020 |access-date=13 March 2020 |vauthors=Oberholzer M, Febbo P |name-list-format=vanc}}</ref> |
|||
<ref name="gisaid">{{Cite web |url=https://platform.gisaid.org/epi3/start/CoV2020 |title=CoV2020 |website=GISAID EpifluDB |url-access=registration |url-status=live |archive-url=https://web.archive.org/web/20200112130540/https://platform.gisaid.org/epi3/start/CoV2020 |archive-date=12 January 2020 |access-date=12 January 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="GuardianLeap">{{Cite news |url=https://www.theguardian.com/world/2020/jan/30/coronavirus-deaths-leap-in-china-as-countries-struggle-to-evacuate-citizens |title=Coronavirus deaths leap in China as countries struggle to evacuate citizens |date=30 January 2020 |work=[[The Guardian]] |access-date=10 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200206150542/https://www.theguardian.com/world/2020/jan/30/coronavirus-deaths-leap-in-china-as-countries-struggle-to-evacuate-citizens |archive-date=6 February 2020 |vauthors=Boseley S, McCurry J |name-list-format=vanc}}</ref> |
|||
<ref name="GZM-20200220">{{Cite web |url=https://gizmodo.com/scientists-create-atomic-level-image-of-the-new-coronav-1841795715 |title=Scientists Create Atomic-Level Image of the New Coronavirus's Potential Achilles Heel |date=19 February 2020 |website=[[Gizmodo]] |url-status=live |archive-url=https://web.archive.org/web/20200308070019/https://gizmodo.com/scientists-create-atomic-level-image-of-the-new-coronav-1841795715 |archive-date=8 March 2020 |access-date=13 March 2020 |vauthors=Mandelbaum RF |name-list-format=vanc}}</ref> |
|||
<ref name="HillWHOname">{{cite news|last1=Gstalter|first1=Morgan|url=https://thehill.com/homenews/administration/488479-who-official-warns-against-calling-it-chinese-virus-says-there-is-no|title=WHO official warns against calling it 'Chinese virus', says 'there is no blame in this'|date=19 March 2020|work=[[The Hill (newspaper)|The Hill]]|accessdate=21 March 2020|archive-url=https://web.archive.org/web/20200418035520/https://thehill.com/homenews/administration/488479-who-official-warns-against-calling-it-chinese-virus-says-there-is-no|archive-date=18 April 2020|url-status=live}}</ref> |
|||
<ref name="HoffmanCell">{{Cite journal |displayauthors=6 |vauthors=Hoffman M, Kliene-Weber H, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S |date=16 April 2020 |title=SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor |journal=[[Cell (journal)|Cell]] |volume=181 |issue=2 |pages=271–280.e8 |doi=10.1016/j.cell.2020.02.052 |pmc=7102627 |pmid=32142651 |name-list-format=vanc}}</ref> |
|||
<ref name="HuangNPR">{{Cite web |url=https://www.npr.org/sections/goatsandsoda/2020/01/22/798277557/how-does-wuhan-coronavirus-compare-to-mers-sars-and-the-common-cold |title=How Does Wuhan Coronavirus Compare with MERS, SARS and the Common Cold? |date=22 January 2020 |website=[[NPR]] |url-status=live |archive-url=https://web.archive.org/web/20200202094021/https://www.npr.org/sections/goatsandsoda/2020/01/22/798277557/how-does-wuhan-coronavirus-compare-to-mers-sars-and-the-common-cold |archive-date=2 February 2020 |access-date=3 February 2020 |vauthors=Huang P |name-list-format=vanc}}</ref> |
|||
<ref name="IJID-interm-host">{{Cite journal |vauthors=Wu D, Wu T, Liu Q, Yang Z |date=12 March 2020 |title=The SARS-CoV-2 outbreak: what we know |url=https://www.ijidonline.com/article/S1201-9712(20)30123-5/fulltext |journal=International Journal of Infectious Diseases |language=English |volume=94 |pages=44–48 |doi=10.1016/j.ijid.2020.03.004 |issn=1201-9712 |pmc=7102543 |pmid=32171952 |name-list-format=vanc |access-date=16 April 2020 |archive-url=https://web.archive.org/web/20200409201324/https://www.ijidonline.com/article/S1201-9712(20%2930123-5/fulltext |archive-date=9 April 2020 |url-status=live}}</ref> |
|||
<ref name="Isolation">{{Cite journal |vauthors=Xiao K, Zhai J, Feng Y |s2cid=213920763 |date=February 2020 |title=Isolation and Characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins |journal=[[bioRxiv]] |type=preprint |doi=10.1101/2020.02.17.951335 |name-list-format=vanc |url=https://www.biorxiv.org/content/biorxiv/early/2020/02/20/2020.02.17.951335.full.pdf |access-date=5 May 2020 |archive-url=https://web.archive.org/web/20200422163506/https://www.biorxiv.org/content/biorxiv/early/2020/02/20/2020.02.17.951335.full.pdf |archive-date=22 April 2020 |url-status=live}}</ref> |
|||
<ref name="Kessler17Apr2020">{{Cite news |last=Kessler |first=Glenn |url=https://www.washingtonpost.com/politics/2020/04/17/trumps-false-claim-that-who-said-coronavirus-was-not-communicable/ |title=Trump's false claim that the WHO said the coronavirus was 'not communicable' |date=17 April 2020 |work=The Washington Post |access-date=17 April 2020 |archive-url=https://archive.today/20200417193804/https://www.washingtonpost.com/politics/2020/04/17/trumps-false-claim-that-who-said-coronavirus-was-not-communicable/ |archive-date=17 April 2020}}</ref> |
|||
<ref name="Kuo21Jan2020">{{Cite news |last=Kuo |first=Lily |url=https://www.theguardian.com/world/2020/jan/20/coronavirus-spreads-to-beijing-as-china-confirms-new-cases |title=China confirms human-to-human transmission of coronavirus |date=21 January 2020 |work=[[The Guardian]] |access-date=18 April 2020 |archive-url=https://web.archive.org/web/20200322001315/https://www.theguardian.com/world/2020/jan/20/coronavirus-spreads-to-beijing-as-china-confirms-new-cases |archive-date=22 March 2020 |url-status=live}}</ref> |
|||
<ref name="LancetBinding">{{Cite journal |displayauthors=6 |vauthors=Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W |date=February 2020 |title=Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding |journal=[[The Lancet]] |volume=395 |issue=10224 |pages=565–574 |doi=10.1016/S0140-6736(20)30251-8 |pmc=7159086 |pmid=32007145 |name-list-format=vanc}}</ref> |
|||
<ref name="LancetCharacteristics">{{Cite journal |displayauthors=6 |vauthors=Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Sia J, You T, Zhang X, Zhang L |date=15 February 2020 |title=Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study |url=https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30211-7/fulltext |url-status=live |journal=[[The Lancet]] |volume=395 |issue=10223 |pages=507–513 |doi=10.1016/S0140-6736(20)30211-7 |pmc=7135076 |pmid=32007143 |archive-url=https://web.archive.org/web/20200131002254/https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30211-7/fulltext |archive-date=31 January 2020 |access-date=9 March 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="LancetClinical">{{Cite journal |displayauthors=6 |vauthors=Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Can B |date=15 February 2020 |title=Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China |url=https://www.thelancet.com/action/showPdf?pii=S0140-6736%2820%2930183-5 |url-status=live |journal=[[The Lancet]] |volume=395 |issue=10223 |pages=497–506 |doi=10.1016/S0140-6736(20)30183-5 |pmc=7159299 |pmid=31986264 |archive-url=https://web.archive.org/web/20200131005857/https://www.thelancet.com/action/showPdf?pii=S0140-6736%2820%2930183-5 |archive-date=31 January 2020 |access-date=26 March 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="LancetLoad">{{Cite journal |last1=To |first1=Kelvin Kai-Wang |last2=Tsang |first2=Owen Tak-Yin |last3=Leung |first3=Wai-Shing |last4=Tam |first4=Anthony Raymond |last5=Wu |first5=Tak-Chiu |last6=Lung |first6=David Christopher |last7=Yip |first7=Cyril Chik-Yan |last8=Cai |first8=Jian-Piao |last9=Chan |first9=Jacky Man-Chun |last10=Chik |first10=Thomas Shiu-Hong |last11=Lau |first11=Daphne Pui-Ling |displayauthors=6 |date=March 2020 |title=Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study |url=https://www.thelancet.com/journals/laninf/article/PIIS1473-3099%2820%2930196-1/fulltext |journal=The Lancet Infectious Diseases |doi=10.1016/S1473-3099(20)30196-1 |access-date=21 April 2020 |last12=Choi |first12=Chris Yau-Chung |last13=Chen |first13=Lin-Lei |last14=Chan |first14=Wan-Mui |last15=Chan |first15=Kwok-Hung |last16=Ip |first16=Jonathan Daniel |last17=Ng |first17=Anthony Chin-Ki |last18=Poon |first18=Rosana Wing-Shan |last19=Luo |first19=Cui-Ting |last20=Cheng |first20=Vincent Chi-Chung |last21=Chan |first21=Jasper Fuk-Woo |last22=Hung |first22=Ivan Fan-Ngai |last23=Chen |first23=Zhiwei |last24=Chen |first24=Honglin |last25=Yuen |first25=Kwok-Yung |volume=20 |issue=5 |pages=565–574 |pmid=32213337 |name-list-format=vanc |pmc=7158907 |archive-url=https://web.archive.org/web/20200417115822/https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30196-1/fulltext |archive-date=17 April 2020 |url-status=live}}</ref> |
|||
<ref name="LancetNowcasting">{{Cite journal |vauthors=Perlman S |date=February 2020 |title=Another Decade, Another Coronavirus |journal=[[The New England Journal of Medicine]] |volume=382 |issue=8 |pages=760–762 |doi=10.1056/NEJMe2001126 |pmc=7121143 |pmid=31978944 |name-list-format=vanc}}</ref> |
|||
<ref name="Letko22Jan2020">{{Cite journal |vauthors=Letko M, Munster V |date=January 2020 |title=Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV |journal=[[bioRxiv]] |type=preprint |doi=10.1101/2020.01.22.915660 |pmid=32511294 |pmc=7217099 |name-list-format=vanc |url=https://www.biorxiv.org/content/biorxiv/early/2020/01/22/2020.01.22.915660.full.pdf |access-date=5 May 2020 |archive-url=https://web.archive.org/web/20200422162807/https://www.biorxiv.org/content/biorxiv/early/2020/01/22/2020.01.22.915660.full.pdf |archive-date=22 April 2020 |url-status=live}}</ref> |
|||
<ref name="LiMar2020">{{Cite journal |last1=Li |first1=Jin-Yan |last2=You |first2=Zhi |last3=Wang |first3=Qiong |last4=Zhou |first4=Zhi-Jian |last5=Qiu |first5=Ye |last6=Luo |first6=Rui |last7=Ge |first7=Xing-Yi |displayauthors=6 |date=March 2020 |title=The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future |url=https://www.sciencedirect.com/science/article/pii/S1286457920300307 |journal=Microbes and Infection |volume=22 |issue=2 |pages=80–85 |doi=10.1016/j.micinf.2020.02.002 |pmid=32087334 |access-date=19 April 2020 |name-list-format=vanc |pmc=7079563 |archive-url=https://web.archive.org/web/20200414061718/https://www.sciencedirect.com/science/article/pii/S1286457920300307 |archive-date=14 April 2020 |url-status=live}}</ref> |
|||
<ref name="MedVirEvolution">{{Cite journal |vauthors=Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M |date=April 2020 |title=The 2019-new coronavirus epidemic: Evidence for virus evolution |journal=[[Journal of Medical Virology]] |volume=92 |issue=4 |pages=455–459 |doi=10.1002/jmv.25688 |pmid=31994738 |name-list-format=vanc|pmc=7166400}}</ref> |
|||
<ref name="NatGeoSoap">{{Cite web |url=https://www.nationalgeographic.com/science/2020/03/why-soap-preferable-bleach-fight-against-coronavirus/ |title=Why soap is preferable to bleach in the fight against coronavirus |date=18 March 2020 |website=[[National Geographic]] |url-status=live |archive-url=https://web.archive.org/web/20200402001042/https://www.nationalgeographic.com/science/2020/03/why-soap-preferable-bleach-fight-against-coronavirus/ |archive-date=2 April 2020 |access-date=2 April 2020 |vauthors=Gibbens S |name-list-format=vanc}}</ref> |
|||
<ref name="NatMicLetko">{{Cite journal |vauthors=Letko M, Marzi A, Munster V |date=February 2020 |title=Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses |journal=[[Nature Microbiology]] |volume=5 |issue=4 |pages=562–569 |doi=10.1038/s41564-020-0688-y |pmc=7095430 |pmid=32094589 |name-list-format=vanc}}</ref> |
|||
<ref name="nature feb2020">{{Cite journal |vauthors=Cyranoski D |date=26 February 2020 |title=Mystery deepens over animal source of coronavirus |journal=[[Nature (journal)|Nature]] |volume=579 |issue=7797 |pages=18–19 |bibcode=2020Natur.579...18C |doi=10.1038/d41586-020-00548-w |pmid=32127703 |name-list-format=vanc|doi-access=free}}</ref> |
|||
<ref name="NaturePang">{{Cite journal |vauthors=Cyranoski D |s2cid=212825975 |date=7 February 2020 |title=Did pangolins spread the China coronavirus to people? |url=https://www.nature.com/articles/d41586-020-00364-2 |url-status=live |journal=[[Nature (journal)|Nature]] |doi=10.1038/d41586-020-00364-2 |archive-url=https://web.archive.org/web/20200207163538/https://www.nature.com/articles/d41586-020-00364-2 |archive-date=7 February 2020 |access-date=12 February 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="NaturePeakLoad">{{Cite journal |displayauthors=6 |vauthors=Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C |date=April 2020 |title=Virological assessment of hospitalized patients with COVID-2019 |journal=Nature |volume=581 |issue=7809 |pages=465–469 |doi=10.1038/s41586-020-2196-x |pmid=32235945|bibcode=2020Natur.581..465W |doi-access=free}}</ref> |
|||
<ref name="NatureTempShedding">{{Cite journal |last1=He |first1=Xi |last2=Lau |first2=Eric H. Y. |last3=Wu |first3=Peng |last4=Deng |first4=Xilong |last5=Wang |first5=Jian |last6=Hao |first6=Xinxin |last7=Lau |first7=Yiu Chung |last8=Wong |first8=Jessica Y. |last9=Guan |first9=Yujuan |last10=Tan |first10=Xinghua |last11=Mo |first11=Xiaoneng |displayauthors=6 |date=15 April 2020 |title=Temporal dynamics in viral shedding and transmissibility of COVID-19 |url=https://www.nature.com/articles/s41591-020-0869-5 |journal=Nature Medicine |doi=10.1038/s41591-020-0869-5 |access-date=21 April 2020 |last12=Chen |first12=Yanqing |last13=Liao |first13=Baolin |last14=Chen |first14=Weilie |last15=Hu |first15=Fengyu |last16=Zhang |first16=Qing |last17=Zhong |first17=Mingqiu |last18=Wu |first18=Yanrong |last19=Zhao |first19=Lingzhai |last20=Zhang |first20=Fuchun |last21=Cowling |first21=Benjamin J. |last22=Li |first22=Fang |last23=Leung |first23=Gabriel M. |volume=26 |issue=5 |pages=672–675 |pmid=32296168 |name-list-format=vanc |archive-url=https://web.archive.org/web/20200419181313/https://www.nature.com/articles/s41591-020-0869-5 |archive-date=19 April 2020 |url-status=live |doi-access=free}}</ref> |
|||
<ref name="NatureZhou">{{Cite journal |displayauthors=6 |vauthors=Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL |date=February 2020 |title=A pneumonia outbreak associated with a new coronavirus of probable bat origin |journal=[[Nature (journal)|Nature]] |volume=579 |issue=7798 |pages=270–273 |doi=10.1038/s41586-020-2012-7 |pmc=7095418 |pmid=32015507 |bibcode=2020Natur.579..270Z |name-list-format=vanc}}</ref> |
|||
<ref name="NBC-GOP">{{Cite news |url=https://www.nbcnews.com/news/asian-america/cdc-chief-spurns-term-chinese-coronavirus-used-gop-lawmakers-n1156656 |title=GOP lawmakers continue to use 'Wuhan virus' or 'Chinese coronavirus' |date=12 March 2020 |access-date=19 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200314185524/https://www.nbcnews.com/news/asian-america/cdc-chief-spurns-term-chinese-coronavirus-used-gop-lawmakers-n1156656 |archive-date=14 March 2020 |publisher=[[NBC News]] |vauthors=Yam K |name-list-format=vanc}}</ref> |
|||
<ref name="NBCSpread">{{Cite news |url=https://www.nbcnews.com/health/health-news/how-does-new-coronavirus-spread-n1121856 |title=How does coronavirus spread? |date=25 January 2020 |access-date=13 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200128081650/https://www.nbcnews.com/health/health-news/how-does-new-coronavirus-spread-n1121856 |archive-date=28 January 2020 |publisher=[[NBC News]] |vauthors=Edwards E |name-list-format=vanc}}</ref> |
|||
<ref name="NCBI-Bat">{{Cite journal |date=15 February 2020 |title=Bat SARS-like coronavirus isolate bat-SL-CoVZC45, complete genome |url=https://www.ncbi.nlm.nih.gov/nuccore/MG772933 |access-date=15 February 2020 |website=[[National Center for Biotechnology Information]] (NCBI) |name-list-format=vanc |archive-url=https://web.archive.org/web/20200604011749/https://www.ncbi.nlm.nih.gov/nuccore/MG772933 |archive-date=4 June 2020 |url-status=live }}</ref> |
|||
<ref name="NCBI-Bat2">{{Cite journal |date=15 February 2020 |title=Bat SARS-like coronavirus isolate bat-SL-CoVZXC21, complete genome |url=https://www.ncbi.nlm.nih.gov/nuccore/MG772934 |access-date=15 February 2020 |website=[[National Center for Biotechnology Information]] (NCBI) |name-list-format=vanc |archive-url=https://web.archive.org/web/20200604011744/https://www.ncbi.nlm.nih.gov/nuccore/MG772934 |archive-date=4 June 2020 |url-status=live }}</ref> |
|||
<ref name="NCBI-Bat3">{{Cite web |url=https://www.ncbi.nlm.nih.gov/nuccore/1802633852 |title=Bat coronavirus isolate RaTG13, complete genome |date=10 February 2020 |website=[[National Center for Biotechnology Information]] (NCBI) |access-date=5 March 2020 |name-list-format=vanc |archive-url=https://web.archive.org/web/20200515133838/https://www.ncbi.nlm.nih.gov/nuccore/1802633852 |archive-date=15 May 2020 |url-status=live}}</ref> |
|||
<ref name="NEJM-Dynamics">{{Cite journal |displayauthors=6 |vauthors=Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Li M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TT, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z |date=January 2020 |title=Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia |journal=[[The New England Journal of Medicine]] |volume=382 |issue=13 |pages=1199–1207 |doi=10.1056/NEJMoa2001316 |pmc=7121484 |pmid=31995857 |name-list-format=vanc}}</ref> |
|||
<ref name="NEJM-FirstUS">{{Cite journal |displayauthors=6 |vauthors=Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK |date=March 2020 |title=First Case of 2019 Novel Coronavirus in the United States |journal=[[The New England Journal of Medicine]] |volume=382 |issue=10 |pages=929–936 |doi=10.1056/NEJMoa2001191 |pmc=7092802 |pmid=32004427 |name-list-format=vanc}}</ref> |
|||
<ref name="NEJM-Novel">{{Cite journal |displayauthors=6 |vauthors=Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W |date=February 2020 |title=A Novel Coronavirus from Patients with Pneumonia in China, 2019 |journal=[[The New England Journal of Medicine]] |volume=382 |issue=8 |pages=727–733 |doi=10.1056/NEJMoa2001017 |pmc=7092803 |pmid=31978945 |name-list-format=vanc}}</ref> |
|||
<ref name="NEJM-Stability">{{Cite journal |displayauthors=6 |vauthors=van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ |date=17 March 2020 |title=Correspondence: Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1 |journal=[[The New England Journal of Medicine]] |volume=382 |issue=16 |pages=1564–1567 |doi=10.1056/NEJMc2004973 |pmc=7121658 |pmid=32182409 |name-list-format=vanc}}</ref> |
|||
<ref name="NexstrainApril">{{Cite web|title=Genomic epidemiology of novel coronavirus - Global subsampling|url=https://nextstrain.org/ncov/global|last=|first=|date=|website=Nextstrain|url-status=live|archive-url=https://web.archive.org/web/20200420123520/https://nextstrain.org/ncov/global|archive-date=20 April 2020|access-date=7 May 2020}}</ref> |
|||
<ref name="NextstrainJanuary">{{Cite web |url=https://nextstrain.org/narratives/ncov/sit-rep/2020-01-30 |title=Genomic analysis of nCoV spread: Situation report 2020-01-30 |website=nextstrain.org |url-status=live <!--Errors loading archive page-->|archive-url=https://web.archive.org/web/20200315182810/https://nextstrain.org/narratives/ncov/sit-rep/2020-01-30 |archive-date=15 March 2020 |access-date=18 March 2020 |vauthors=Bedford T, Neher R, Hadfield N, Hodcroft E, Ilcisin M, Müller N}}</ref> |
|||
<ref name="NextstrainPhylogeny">{{Cite web |url=https://nextstrain.org/groups/blab/sars-like-cov |title=Phylogeny of SARS-like betacoronaviruses |website=nextstrain |url-status=live |archive-url=https://web.archive.org/web/20200120190511/https://nextstrain.org/groups/blab/sars-like-cov |archive-date=20 January 2020 |access-date=18 January 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="NG-20200415">{{cite news |last=Wu |first=Katherine J. |title=There are more viruses than stars in the universe. Why do only some infect us? - More than a quadrillion quadrillion individual viruses exist on Earth, but most are not poised to hop into humans. Can we find the ones that are? |url=https://www.nationalgeographic.com/science/2020/04/factors-allow-viruses-infect-humans-coronavirus/ |date=15 April 2020 |work=[[National Geographic Society]] |accessdate=18 May 2020 |archive-url=https://web.archive.org/web/20200423000758/https://www.nationalgeographic.com/science/2020/04/factors-allow-viruses-infect-humans-coronavirus/ |archive-date=23 April 2020 |url-status=live}}</ref> |
|||
<ref name="NIH-Structure">{{Cite web |url=https://www.nih.gov/news-events/nih-research-matters/novel-coronavirus-structure-reveals-targets-vaccines-treatments |title=Novel coronavirus structure reveals targets for vaccines and treatments |date=2 March 2020 |website=[[National Institutes of Health]] (NIH) |url-status=live |archive-url=https://web.archive.org/web/20200401205344/https://www.nih.gov/news-events/nih-research-matters/novel-coronavirus-structure-reveals-targets-vaccines-treatments |archive-date=1 April 2020 |access-date=3 April 2020}}</ref> |
|||
<ref name="nihSARSr-CoV">{{cite web |title=New coronavirus stable for hours on surfaces |url=https://www.nih.gov/news-events/news-releases/new-coronavirus-stable-hours-surfaces |website=National Institutes of Health (NIH) |publisher=NIH.gov |accessdate=4 May 2020 |language=EN |date=17 March 2020 |archive-url=https://web.archive.org/web/20200323032520/https://www.nih.gov/news-events/news-releases/new-coronavirus-stable-hours-surfaces |archive-date=23 March 2020 |url-status=live}}</ref> |
|||
<ref name="November case">{{Cite news |url=https://www.scmp.com/news/china/society/article/3074991/coronavirus-chinas-first-confirmed-covid-19-case-traced-back |title=Coronavirus: China's first confirmed Covid-19 case traced back to November 17 |date=13 March 2020 |access-date=16 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200313004217/https://www.scmp.com/news/china/society/article/3074991/coronavirus-chinas-first-confirmed-covid-19-case-traced-back |archive-date=13 March 2020 |website=[[South China Morning Post]] |vauthors=Ma J |name-list-format=vanc}}</ref> |
|||
<ref name="NYT 20200130">{{Cite news |url=https://www.nytimes.com/2020/01/30/health/coronavirus-world-health-organization.html |title=W.H.O. Declares Global Emergency as Wuhan Coronavirus Spreads |date=30 January 2020 |access-date=30 January 2020 |url-status=live |archive-url=https://web.archive.org/web/20200130195011/https://www.nytimes.com/2020/01/30/health/coronavirus-world-health-organization.html |archive-date=30 January 2020 |vauthors=Wee SL, ((McNeil Jr. DG)), Hernández JC |website=[[The New York Times]] |name-list-format=vanc}}</ref> |
|||
<ref name="NYT-Ban">{{Cite news |url=https://www.nytimes.com/2020/02/27/science/coronavirus-pangolin-wildlife-ban-china.html |title=China's Ban on Wildlife Trade a Big Step, but Has Loopholes, Conservationists Say |date=27 February 2020 |work=[[The New York Times]] |access-date=23 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200313060353/https://www.nytimes.com/2020/02/27/science/coronavirus-pangolin-wildlife-ban-china.html |archive-date=13 March 2020 |vauthors=Gorman J |name-list-format=vanc}}</ref> |
|||
<ref name="NYT-SpikyBlob">{{Cite news |url=https://www.nytimes.com/2020/04/01/health/coronavirus-illustration-cdc.html |title=The Spiky Blob Seen Around the World |date=1 April 2020 |work=[[The New York Times]] |access-date=6 April 2020 |vauthors=Giaimo C |name-list-format=vanc |archive-url=https://web.archive.org/web/20200402012708/https://www.nytimes.com/2020/04/01/health/coronavirus-illustration-cdc.html |archive-date=2 April 2020 |url-status=live}}</ref> |
|||
<ref name="NYT6">{{Cite news |url=https://www.nytimes.com/2020/03/04/us/coronavirus-recovery.html |title=We Spoke to Six Americans with Coronavirus |date=4 March 2020 |access-date=16 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200313191200/https://www.nytimes.com/2020/03/04/us/coronavirus-recovery.html |archive-date=13 March 2020 |website=[[The New York Times]] |vauthors=Harmon A |name-list-format=vanc}}</ref> |
|||
<ref name="oie.int">{{Cite web |url=https://www.oie.int/en/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/ |title=Questions and Answers on the COVID-19: OIE - World Organisation for Animal Health |website=www.oie.int |access-date=16 April 2020 |archive-url=https://web.archive.org/web/20200331185802/https://www.oie.int/en/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/ |archive-date=31 March 2020 |url-status=live}}</ref> |
|||
<ref name="pmid32014114">{{Cite journal |vauthors=Wu JT, Leung K, Leung GM |date=February 2020 |title=Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study |journal=[[The Lancet]] |volume=395 |issue=10225 |pages=689–697 |doi=10.1016/S0140-6736(20)30260-9 |pmc=7159271 |pmid=32014114 |name-list-format=vanc}}</ref> |
|||
<ref name="PNAS2020">{{Cite journal |vauthors=Forster P, Forster L, Renfrew C, Forster M |date=8 April 2020 |title=Phylogenetic network analysis of SARS-CoV-2 genomes |url=https://www.pnas.org/content/pnas/early/2020/04/07/2004999117.full.pdf |journal=PNAS |volume=117 |issue=17 |pages=9241–9243 |doi=10.1073/pnas.2004999117 |pmid=32269081 |pmc=7196762 |language=en |access-date=17 April 2020 |name-list-format=vanc |archive-url=https://web.archive.org/web/20200416234149/https://www.pnas.org/content/pnas/early/2020/04/07/2004999117.full.pdf |archive-date=16 April 2020 |url-status=live}}</ref> |
|||
<ref name="PopSciJan">{{Cite web |url=https://www.popsci.com/story/health/wuhan-coronavirus-china-wet-market-wild-animal/ |title=We're still not sure where the Wuhan coronavirus really came from |date=28 January 2020 |website=[[Popular Science]] |url-status=live |archive-url=https://archive.today/20200130003350/https://www.popsci.com/story/health/wuhan-coronavirus-china-wet-market-wild-animal/ |archive-date=30 January 2020 |access-date=30 January 2020 |vauthors=Eschner K |name-list-format=vanc}}</ref> |
|||
<ref name="Proximal">{{Cite journal |vauthors=Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF |date=17 March 2020 |title=Correspondence: The proximal origin of SARS-CoV-2 |journal=[[Nature Medicine]] |volume=26 |issue=4 |pages=450–452 |doi=10.1038/s41591-020-0820-9 |pmc=7095063 |pmid=32284615 |name-list-format=vanc}}</ref> |
|||
<ref name="QZ-Why">{{Cite news |url=https://qz.com/1820422/coronavirus-why-wont-who-use-the-name-sars-cov-2/ |title=Why won't the WHO call the coronavirus by its name, SARS-CoV-2? |date=18 March 2020 |work=[[Quartz (publication)|Quartz]] |access-date=26 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200325235531/https://qz.com/1820422/coronavirus-why-wont-who-use-the-name-sars-cov-2/ |archive-date=25 March 2020 |vauthors=Hui M |name-list-format=vanc}}</ref> |
|||
<ref name="Rejects">{{Cite journal |vauthors=Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S |date=April 2020 |title=Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event |url=https://www.sciencedirect.com/science/article/pii/S1567134820300447 |journal=[[Infection, Genetics and Evolution]] |volume=79 |pages=104212 |doi=10.1016/j.meegid.2020.104212 |pmc=7106301 |pmid=32004758 |access-date=9 April 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="Rocklov">{{Cite journal |vauthors=Rocklöv J, Sjödin H, Wilder-Smith A |date=February 2020 |title=COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures |journal=Journal of Travel Medicine |volume=27 |issue=3 |doi=10.1093/jtm/taaa030 |pmc=7107563 |pmid=32109273 |name-list-format=vanc}}</ref> |
|||
<ref name="SCI-20200219">{{Cite journal |displayauthors=6 |vauthors=Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS |date=February 2020 |title=Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation |journal=[[Science (journal)|Science]] |volume=367 |issue=6483 |pages=1260–1263 |bibcode=2020Sci...367.1260W |doi=10.1126/science.abb2507 |pmid=32075877 |name-list-format=vanc|pmc=7164637}}</ref> |
|||
<ref name="ScienceFlawed">{{Cite journal |vauthors=Kupferschmidt K |date=February 2020 |title=Study claiming new coronavirus can be transmitted by people without symptoms was flawed |journal=[[Science (journal)|Science]] |doi=10.1126/science.abb1524 |name-list-format=vanc}}</ref> |
|||
<ref name="ScienceRapid">{{Cite journal |displayauthors=6 |vauthors=Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J |date=16 March 2020 |title=Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) |journal=[[Science (journal)|Science]] |volume=368 |issue=6490 |pages=489–493 |doi=10.1126/science.abb3221 |pmid=32179701 |name-list-format=vanc|pmc=7164387|bibcode=2020Sci...368..489L }}</ref> |
|||
<ref name="SCLSModeling">{{Cite journal |displayauthors=6 |vauthors=Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P |date=March 2020 |title=Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission |journal=[[Science China Life Sciences]] |volume=63 |issue=3 |pages=457–460 |doi=10.1007/s11427-020-1637-5 |pmc=7089049 |pmid=32009228 |name-list-format=vanc}}</ref> |
|||
<!-- <ref name="SCMP24March2020">[https://www.scmp.com/news/world/united-states-canada/article/3076522/donald-trump-drops-chinese-virus-terminology-white "Donald Trump drops 'Chinese virus' terminology in White House briefing, calls for protecting Asian-Americans"] {{Webarchive|url=https://web.archive.org/web/20200421165146/https://www.scmp.com/news/world/united-states-canada/article/3076522/donald-trump-drops-chinese-virus-terminology-white |date=21 April 2020}}, ''South China Morning Post'', 24 March 2020</ref>--> |
|||
<ref name="Semen">{{cite journal |last1=Li |first1=Diangeng |last2=Jin |first2=Meiling |last3=Bao |first3=Pengtao |last4=Zhao |first4=Weiguo |last5=Zhang |first5=Shixi |title=Clinical Characteristics and Results of Semen Tests Among Men With Coronavirus Disease 2019 |journal=JAMA Network Open |date=7 May 2020 |volume=3 |issue=5 |pages=e208292 |doi=10.1001/jamanetworkopen.2020.8292 |pmid=32379329 |pmc=7206502 |name-list-format=vanc}}</ref> |
|||
<ref name="STAT-Severity">{{Cite news |url=https://www.statnews.com/2020/01/30/limited-data-may-skew-assumptions-severity-coronavirus-outbreak/ |title=Limited data on coronavirus may be skewing assumptions about severity |date=30 January 2020 |access-date=13 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200201161634/https://www.statnews.com/2020/01/30/limited-data-may-skew-assumptions-severity-coronavirus-outbreak/ |archive-date=1 February 2020 |publisher=[[Stat (website)|STAT]] |vauthors=Branswell H |name-list-format=vanc}}</ref> |
|||
<ref name="SunChinaAfrica">{{Cite news |url=https://www.sunnewsonline.com/coronavirus-china-to-repay-africa-in-safeguarding-public-health/ |title=Coronavirus: China to repay Africa in safeguarding public health |date=25 February 2020 |work=[[The Sun (Nigeria)|The Sun]] |access-date=10 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200309103243/https://www.sunnewsonline.com/coronavirus-china-to-repay-africa-in-safeguarding-public-health/ |archive-date=9 March 2020 |vauthors=Paulinus A |name-list-format=vanc}}</ref> |
|||
<ref name="TelegraphPangolins">{{Cite news |url=https://www.telegraph.co.uk/science/2016/03/15/pangolins-13-facts-about-the-worlds-most-hunted-animal/ |title=Pangolins: 13 facts about the world's most hunted animal |date=1 January 2015 |work=The Telegraph |access-date=9 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20191224153802/https://www.telegraph.co.uk/science/2016/03/15/pangolins-13-facts-about-the-worlds-most-hunted-animal/ |archive-date=24 December 2019 |vauthors=Kelly G |name-list-format=vanc}}</ref> |
|||
<ref name="Thorax">''[[Daily Telegraph]]'', Thursday 28 May 2020, page 2 column 1, which refers to the medical journal [[Thorax (journal)|''Thorax'']]; ''Thorax'' May 2020 article [https://thorax.bmj.com/content/early/2020/05/27/thoraxjnl-2020-215091 ''COVID-19: in the footsteps of Ernest Shackleton''] {{Webarchive|url=https://web.archive.org/web/20200530144907/https://thorax.bmj.com/content/early/2020/05/27/thoraxjnl-2020-215091 |date=30 May 2020 }}</ref> |
|||
<ref name="TodayNameMixup">{{cite news |title=Novel coronavirus named 'Covid-19': WHO |url=https://www.todayonline.com/world/wuhan-novel-coronavirus-named-covid-19-who |access-date=11 February 2020 |publisher=TODAYonline |name-list-format=vanc |archive-url=https://archive.today/20200321085608/https://www.todayonline.com/world/wuhan-novel-coronavirus-named-covid-19-who |archive-date=21 March 2020 |url-status=live}}</ref> |
|||
<ref name="USDAtiger">{{Cite web |url=https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/ny-zoo-covid-19 |title=USDA Statement on the Confirmation of COVID-19 in a Tiger in New York |date=5 April 2020 |website=[[United States Department of Agriculture]] |url-status=live |access-date=16 April 2020 |archive-url=https://web.archive.org/web/20200415155712/https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/ny-zoo-covid-19 |archive-date=15 April 2020}}</ref> |
|||
<ref name="USnews17March2020">{{cite news|last1=Shinkman|first1=Paul|url=https://www.usnews.com/news/national-news/articles/2020-03-17/trump-refutes-claims-of-anti-chinese-stigma-in-coronavirus-reponse|title=Trump Fires Back at Complaints He's Stigmatizing China Over Coronavirus|date=17 March 2020|work=[[US News]]|accessdate=21 March 2020|archive-url=https://web.archive.org/web/20200329175146/https://www.usnews.com/news/national-news/articles/2020-03-17/trump-refutes-claims-of-anti-chinese-stigma-in-coronavirus-reponse|archive-date=29 March 2020|url-status=live}}</ref> |
|||
<ref name="VirologicalInitial">{{Cite web |url=http://virological.org/t/initial-genome-release-of-novel-coronavirus/319 |title=Initial genome release of novel coronavirus |date=11 January 2020 |website=Virological |url-status=live |archive-url=https://web.archive.org/web/20200112100227/http://virological.org/t/initial-genome-release-of-novel-coronavirus/319 |archive-date=12 January 2020 |access-date=12 January 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="VirusesPangolins">{{Cite journal |vauthors=Liu P, Chen W, Chen JP |date=October 2019 |title=Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica) |journal=[[Viruses (journal)|Viruses]] |volume=11 |issue=11 |page=979 |doi=10.3390/v11110979 |pmc=6893680 |pmid=31652964 |name-list-format=vanc}}</ref> |
|||
<ref name="WHO-PHEIC">{{Cite press release |title=Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) |date=30 January 2020 |url=https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) |access-date=30 January 2020 |archive-url=https://web.archive.org/web/20200131005904/https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) |archive-date=31 January 2020 |website=[[World Health Organization]] (WHO) |url-status=live |name-list-format=vanc}}</ref> |
|||
<ref name="WHO-SR10">{{Cite report |title=Novel Coronavirus (2019-nCoV): situation report, 10 |date=30 January 2020 |publisher=[[World Health Organization]] |hdl-access=free |vauthors=((World Health Organization)) |hdl=10665/330775 |name-list-format=vanc}}</ref> |
|||
<ref name="WHO-SR12">{{Cite report |title=Novel Coronavirus (2019-nCoV): situation report, 12 |date=1 February 2020 |publisher=[[World Health Organization]] |hdl-access=free |vauthors=((World Health Organization)) |hdl=10665/330777 |name-list-format=vanc}}</ref> |
|||
<ref name="WHO-SR22">{{Cite report |title=Novel Coronavirus (2019-nCoV): situation report, 22 |date=11 February 2020 |publisher=[[World Health Organization]] |hdl-access=free |hdl=10665/330991 |name-list-format=vanc}}</ref> |
|||
<ref name="WHO-SR69">{{Cite report |title=Coronavirus disease 2019 (COVID-19) Situation Report – 69 |date=29 March 2020 |publisher=[[World Health Organization]] |hdl-access=free |hdl=10665/331615 |name-list-format=vanc}}</ref> |
|||
<ref name="WHO-Workplace">{{Cite web |url=https://www.who.int/docs/default-source/coronaviruse/getting-workplace-ready-for-covid-19.pdf |title=Getting your workplace ready for COVID-19 |date=27 February 2020 |website=World Health Organization |url-status=live |archive-url=https://web.archive.org/web/20200302092018/https://www.who.int/docs/default-source/coronaviruse/getting-workplace-ready-for-covid-19.pdf |archive-date=2 March 2020 |access-date=3 March 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="WHO2020QA">{{Cite web |url=https://www.who.int/news-room/q-a-detail/q-a-coronaviruses |title=Q&A on coronaviruses (COVID-19) |date=11 February 2020 |website=[[World Health Organization]] (WHO) |url-status=live |archive-url=https://web.archive.org/web/20200120174649/https://www.who.int/news-room/q-a-detail/q-a-coronaviruses |archive-date=20 January 2020 |access-date=24 February 2020 |name-list-format=vanc}}</ref> |
|||
<ref name="WHO21Jan2020">{{Cite report |title=Surveillance case definitions for human infection with novel coronavirus (nCoV): interim guidance v1, January 2020 |date=January 2020 |publisher=World Health Organization |id=WHO/2019-nCoV/Surveillance/v2020.1 |hdl-access=free |hdl=10665/330376 |name-list-format=vanc}}</ref> |
|||
<ref name="WHOChinaJoint">{{Cite report |url=https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf |title=Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) |date=24 February 2020 |publisher=[[World Health Organization]] (WHO) |access-date=5 March 2020 |archive-url=https://web.archive.org/web/20200229221222/https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf |archive-date=29 February 2020 |url-status=live |name-list-format=vanc}}</ref> |
|||
<ref name="WHOnamingguidelines">{{cite web|url=https://apps.who.int/iris/bitstream/handle/10665/163636/WHO_HSE_FOS_15.1_eng.pdf|title=World Health Organization Best Practices for the Naming of New Human Infectious Diseases|last=|first=|date=May 2015|website=WHO|url-status=live|archive-url=https://web.archive.org/web/20200212201906/https://apps.who.int/iris/bitstream/handle/10665/163636/WHO_HSE_FOS_15.1_eng.pdf|archive-date=12 February 2020|access-date=}}</ref> |
|||
<ref name="WHOPandemic">{{Cite press release |title=WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020 |date=11 March 2020 |url=https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 |access-date=12 March 2020 |archive-url=https://web.archive.org/web/20200311212521/https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 |archive-date=11 March 2020 |website=[[World Health Organization]] (WHO) |url-status=live |name-list-format=vanc}}</ref> |
|||
<ref name="Wong2019">{{Cite journal |vauthors=Wong AC, Li X, Lau SK, Woo PC |date=February 2019 |title=Global Epidemiology of Bat Coronaviruses |journal=[[Viruses (journal)|Viruses]] |volume=11 |issue=2 |page=174 |doi=10.3390/v11020174 |pmc=6409556 |pmid=30791586 |name-list-format=vanc}}</ref> |
|||
<ref name="WongRecombination">{{Cite journal |vauthors=Wong MC, Cregeen SJ, Ajami NJ, Petrosino JF |date=February 2020 |title=Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019 |journal=[[bioRxiv]] |type=preprint |doi=10.1101/2020.02.07.939207 |pmid=32511310 |pmc=7217297 |name-list-format=vanc |url=https://www.biorxiv.org/content/biorxiv/early/2020/02/13/2020.02.07.939207.full.pdf |access-date=5 May 2020 |archive-url=https://web.archive.org/web/20200422162708/https://www.biorxiv.org/content/biorxiv/early/2020/02/13/2020.02.07.939207.full.pdf |archive-date=22 April 2020 |url-status=live}}</ref> |
|||
<ref name="WSJPandemic">{{Cite news |url=https://www.wsj.com/articles/u-s-coronavirus-cases-top-1-000-11583917794 |title=Coronavirus Declared Pandemic by World Health Organization |date=11 March 2020 |access-date=12 March 2020 |url-status=live |archive-url=https://web.archive.org/web/20200311225704/https://www.wsj.com/articles/u-s-coronavirus-cases-top-1-000-11583917794 |archive-date=11 March 2020 |vauthors=McKay B, Calfas J, Ansari T |website=[[The Wall Street Journal]] |name-list-format=vanc}}</ref> |
|||
<ref name="WuStructure">{{Cite journal |displayauthors=6 |vauthors=Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H |date=February 2020 |title=Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods |journal=Acta Pharmaceutica Sinica B |volume=10 |issue=5 |pages=766–788 |doi=10.1016/j.apsb.2020.02.008 |pmc=7102550 |pmid=32292689 |name-list-format=vanc}}</ref> |
|||
<ref name="XivDecoding">{{Cite journal |vauthors=Yu WB, Tang GD, Zhang L, Corlett RT |date=21 February 2020 |title=Decoding evolution and transmissions of novel pneumonia coronavirus using the whole genomic data |url=http://www.chinaxiv.org/abs/202002.00033 |url-status=live |doi=10.12074/202002.00033 |doi-broken-date=2020-08-23 |archive-url=https://web.archive.org/web/20200223200715/http://www.chinaxiv.org/abs/202002.00033 |archive-date=23 February 2020 |access-date=25 February 2020 |website=ChinaXiv |name-list-format=vanc}}</ref> |
|||
<ref name="Zoonotic">{{Cite journal |pmid = 32314559|year = 2020 |last1 = Wong|first1 = G.|last2 = Bi|first2 = Y. H.|last3 = Wang|first3 = Q. H.|last4 = Chen|first4 = X. W.|last5 = Zhang|first5 = Z. G.|last6 = Yao|first6 = Y. G.|title = Zoonotic origins of human coronavirus 2019 (HCoV-19 / SARS-CoV-2): Why is this work important?|journal = Zoological Research|volume = 41|issue = 3|pages = 213–219|doi = 10.24272/j.issn.2095-8137.2020.031|pmc = 7231470}}</ref> |
|||
}} |
|||
==Further reading== |
|||
{{Refbegin}} |
|||
* {{Cite journal |last1=Bar-On |first1=Yinon M |last2=Flamholz |first2=Avi |last3=Phillips |first3=Rob |last4=Milo |first4=Ron |date=31 March 2020 |title=SARS-CoV-2 (COVID-19) by the numbers |journal=[[eLife]] |volume=9 |arxiv=2003.12886 |bibcode=2020arXiv200312886B |doi=10.7554/eLife.57309 |pmid=32228860 |pmc=7224694 |name-list-format=vanc |ref=none}} |
|||
* {{Cite journal |last=Brüssow |first=Harald |date=March 2020 |title=The Novel Coronavirus – A Snapshot of Current Knowledge |journal=Microbial Biotechnology |volume=2020 |issue=3 |pages=607–612 |doi=10.1111/1751-7915.13557 |pmc=7111068 |pmid=32144890 |name-list-format=vanc| ref=none}} |
|||
* {{Cite journal |last1=Cascella |first1=Marco |last2=Rajnik |first2=Michael |last3=Cuomo |first3=Arturo |last4=Dulebohn |first4=Scott C. |last5=Di Napoli |first5=Raffaela |date=January 2020 |title=Features, Evaluation and Treatment Coronavirus (COVID-19) |url=https://www.ncbi.nlm.nih.gov/books/NBK554776/ |journal=StatPearls |pmid=32150360 |name-list-format=vanc |ref=none |access-date=4 April 2020 |archive-url=https://web.archive.org/web/20200406135818/https://www.ncbi.nlm.nih.gov/books/NBK554776/ |archive-date=6 April 2020 |url-status=live }} |
|||
* {{Cite journal |last1=Habibzadeh |first1=Parham |last2=Stoneman |first2=Emily K. |date=February 2020 |title=The Novel Coronavirus: A Bird's Eye View |journal=The International Journal of Occupational and Environmental Medicine |volume=11 |issue=2 |pages=65–71 |doi=10.15171/ijoem.2020.1921 |pmid=32020915 |pmc=7205509 |name-list-format=vanc|doi-access=free |ref=none}} |
|||
* {{Cite report |title=Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases |date=2 March 2020 |publisher=[[World Health Organization]] |hdl-access=free |hdl=10665/331329 |name-list-format=vanc |ref=none}} |
|||
{{Refend}} |
|||
==External links== |
|||
{{Scholia|Q82069695}} |
|||
* {{Cite web |url=https://www.cdc.gov/coronavirus/2019-ncov/index.html |title=Coronavirus Disease 2019 (COVID-19) |website=[[Centers for Disease Control and Prevention]] (CDC)|date=11 February 2020 }} |
|||
* {{Cite web |url=https://www.who.int/emergencies/diseases/novel-coronavirus-2019 |title=Coronavirus disease (COVID-19) Pandemic |website=[[World Health Organization]] (WHO)}} |
|||
* {{Cite web |url=https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/ |title=SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) Sequences |website=[[National Center for Biotechnology Information]] (NCBI)}} |
|||
* {{Cite web |url=https://www.thelancet.com/coronavirus |title=COVID-19 Resource Centre |website=[[The Lancet]]}} |
|||
* {{Cite web |url=https://www.nejm.org/coronavirus |title=Coronavirus (Covid-19) |website=[[The New England Journal of Medicine]]}} |
|||
* {{Cite web |url=https://novel-coronavirus.onlinelibrary.wiley.com/ |title=Covid-19: Novel Coronavirus Outbreak |website=[[Wiley (publisher)|Wiley]]}} |
|||
* {{Cite web |url=https://www.viprbrc.org/brc/home.spg?decorator=corona_ncov |title=SARS-CoV-2 |website=[[Virus Pathogen Database and Analysis Resource]]}} |
|||
* {{Cite web |url=http://www.rcsb.org/news?year=2020&article=5e3c4bcba5007a04a313edcc |title=SARS-CoV-2 related protein structures |website=[[Protein Data Bank]]}} |
|||
{{Medical resources| ICD10 = {{ICD10|U07.1}}}} |
|||
{{COVID-19 pandemic}} |
|||
{{Human coronaviruses}} |
|||
{{Viral systemic diseases|state=collapsed}} |
|||
{{Zoonotic viral diseases|state=collapsed}} |
|||
{{subject bar|commons = y|commons-search = Category:SARS-CoV-2|species = y|species-search = |voy = y|voy-search = SARS-CoV-2|n = y|n-search = SARS-CoV-2|wikt = y|wikt-search = |b = y|b-search = SARS-CoV-2|q = y|q-search = SARS-CoV-2|s = y|s-search = "SARS-CoV-2"|v = y|v-search = |d = y|d-search = Q82069695}} |
|||
{{Taxonbar|from=Q82069695}} |
|||
{{Authority control}} |
|||
[[Category:COVID-19]] |
|||
[[Category:Infraspecific virus taxa]] |
|||
[[Category:Sarbecovirus]] |
|||
[[Category:Zoonoses]] |
|||
[[Category:Chiroptera-borne diseases]] |
|||
A lap 2020. augusztus 25., 14:52-kori változata
| Hollófernyiges/próbalap2 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 A SARS-CoV-2 transzmissziós elektronmikroszkópos képe
| ||||||||||||||||
| Vírusbesorolás | ||||||||||||||||
|
A SARS-CoV-2 (az angol Severe acute respiratory syndrome coronavirus 2 rövidítése; magyarul súlyos akut légzőszervi szindróma-koronavírus 2) a Coronaviridae családba tartozó, embereket fertőző vírustörzs, amely a 2019-es koronavírus-betegség (COVID–19) kórokozója.
SARS-CoV-2 a Baltimore-féle osztályozási rendszerben a IV. csoportba (egyszálú, pozitív-szenz RNS-genommal rendelkező vírusok) tartozik, virionját lipidburok veszi körbe.[1][2] Taxonómiai szempontból a SARSr-CoV (súlyos akut légzőszervi szindrómához kapcsolódó koronavírus) faj egyik törzse,[3] akárcsak közeli rokona, a 2002-2004-es SARS világjárványt okozó SARS-CoV-1.[4][5]
A vírus zoonotikus eredetű, genetikai vizsgálatok szerint legközelebbi rokonai a denevérekben élnek.[6][7][8][9] Egyes feltételezések szerint a tobzoskák köztesgazdaként szolgálhattak a denevérek és az emberek között, de ez az elmélet még nincs bizonyítva.[10][11] A vírus genetikai diverzitása alacsony, vagyis az emberre való "átugrása" nemrég, feltehetően 2019 végén következhetett be.[12]
Az epidemiológiai vizsgálatok szerint a vírus a védekező intézkedéseket nem hozó, immunológiailag nem védett közösségekben igen gyorsan terjed, egy beteg 1,4-3,9 másik embernek adja tovább a fertőzést. Terjedését a testi érintkezés vagy a köhögés, tüsszentés vagy akár a beszéd által generált cseppfertőzés biztosítja.[13][14] Gazdasejtjébe az angiotenzin-konvertáló enzim-2 (ACE2) receptorhoz kapcsolódva jut be..[6][15][16][17]
Terjedése
A SARS-CoV-2 emberről emberre való terjedését már a vuhani járvány elején, 2020. január 20-án igazolták.[18][19][20][21] Eleinte úgy vélték, hogy a kórokozó elsősorban a köhögés és tüsszögés kiváltotta cseppfertőzéssel terjed 1,5-2 méteren belül.[14][22] Lézeres fényszóródási vizsgálatokkal azonban kimutatták, hogy a közönséges beszéd is generál apró folyadékcsöppeket, amelyekben a vírus megbújhat;[23][24] sőt a vírusrészecskék magukban is kikerülhetnek a levegőbe.[25]
A leülepedett cseppekkel fertőzött felületek fizikai érintése is veszélyes lehet.[26] A kutatások szerint a SARS-CoV-2 műanyag- és acélfelületeken akár három napig, kartonpapíron egy napig, rézfelületen pedig négy óráig marad életképes.[4] A detergensekkel (mint a szappan) való érintkezés felbontja a vírus külső lipidburkát és inaktiválja azt.[27][28] A vírus RNS-ét kimutatták a beteg különféle testfolyadékaiból, például aspermából, sőt a székletből is.[29][30]
A kórokozó fertőzőképessége a betegség, illetve a korai, tünetmentes szakasz alatt még nem teljesen ismert, de a jelenlegi adatok szerint a torokban a virionszám nagyjából a fertőzés utáni negyedik napon[31][32] vagy tünetek megjelenése utáni első héten a legmagasabb, utána pedig fokozatosan csökken.[33] A WHO első megállapításaival ellentétben[34] az epidemiológiai modellek arra utalnak, hogy a teljesen tünetmentes, illetve korai fázisban lévő betegek az új fertőzések legfőbb forrásai.[35] Egy Montevideóban kikötött óceánjáró 217 utasa és legénysége közül 128-nak lett pozitív a tesztje, míg tüneteket csak 24-en észleltek.[36] Egy 94 betegen elvégzett vizsgálat arra utal, hogy leginkább 2-3 nappal a tünetek megjelenése előtt fertőzőképesek.[37]
Ritka esetekben előfordul, hogy a vírus emberről állatra terjed át, például macskákra,[38][39] emiatt egyes intézmények azt javasolják, hogy a betegek lehetőleg ne érintkezzenek háziállatokkal.[40][41]
Eredete

A SARS-CoV-2-fertőzés első eseteit a kínai Vuhan városában észlelték.[6] Az állatról emberre való átadódás körülményei egyelőre tisztázatlanok.[12][42][9] A betegek közül sokan a vuhani Huanan élelmiszerpiac dolgozói voltak,[43][44] ezért feltételezik, hogy a humán patogén törzs itt alakulhatott ki.[9][45] Nem zárható ki azonban, hogy a piacra is kívülről került a vírus és csak itt kezdett gyors terjedésbe.[12][46] A korai megbetegedésekből (2019 december-2020 február) származó 160 minta alapján a SARS-CoV-2 olyan denevér-koronavírusokhoz hasonlít a leginkább, amelyek Kanton tartományban a leggyakoribbak.[47][48]
A 2002-2004-es SARS-járvány után átfogó kutatás indult a hasonló, állatokban élő vírusok után és kimutatták, hogy számos denevérfaj, elsősorban a parkósorrú denevérek (a Rhinolophus nemzetség tagjai) hordoznak hasonló koronavírusokat. A Rhinolophus sinicus egyik vírusa 80%-os,[8][49][50]míg a Rhinolophus affinis vírustörzse 96%-os hasonlóságot mutatott a SARS-CoV-2-vel.[6][51]

Bats were initially considered to be the most likely natural reservoir of SARS-CoV-2,[52][53] which means that they harbour the virus for long periods of time with no pathogenic effects. Regarding the animal source of infection into humans, the differences between the bat coronavirus sampled at the time and SARS-CoV-2 suggested that humans were infected via an intermediate host. Arinjay Banerjee, a virologist at McMaster University, notes that "the SARS virus shared 99.8% of its genome with a civet coronavirus, which is why civets were considered the source."[45] Although studies had suggested some likely candidates, the number and identities of intermediate hosts remains uncertain.[54] Nearly half of the strain's genome had a phylogenetic lineage distinct from known relatives.[55]

A phylogenetics study published in 2020 indicates that pangolins are a reservoir host of SARS-CoV-2-like coronaviruses.[57] However, there is no direct evidence to link pangolins as an intermediate host of SARS-CoV-2 at this moment. While there is scientific consensus that bats are the ultimate source of coronaviruses, it is hypothesized that a SARS-CoV-2-like coronavirus originated in pangolins, jumped back to bats, and then jumped to humans, resulting in SARS-CoV-2. Based on whole genome sequence similarity, a pangolin coronavirus candidate strain was found to be less similar than RaTG13, but more similar than other bat coronaviruses to SARS-CoV-2.[56] Therefore, based on maximum parsimony, a specific population of bats is more likely to have directly transmitted SARS-CoV-2 to humans than a pangolin, while an evolutionary ancestor to bats was the source of general coronaviruses.[58]
A metagenomics study published in 2019 had previously revealed that SARS-CoV, the strain of the virus that causes SARS, was the most widely distributed coronavirus among a sample of Sunda pangolins.[59] On 7 {{||}}February 2020, South China Agricultural University in Guangzhou announced that researchers discovered a pangolin sample with a particular coronavirus – a single nucleic acid sequence of the virus was "99% similar" to that of a protein-coding RNA of SARS-CoV-2.[60] The authors state that "the receptor-binding domain of the S protein [that binds to the cell surface receptor during infection] of the newly discovered Pangolin-CoV is virtually identical to that of 2019-nCoV, with one amino acid difference."[61] Microbiologists and geneticists in Texas have independently found evidence of reassortment in coronaviruses suggesting involvement of pangolins in the origin of SARS-CoV-2.[62] The majority of the viral RNA is related to a variation of bat coronaviruses. The spike protein appears to be a notable exception, however, possibly acquired through a more recent recombination event with a pangolin coronavirus.[63] Structural analysis of the receptor binding domain (RBD) and human angiotensin-converting enzyme 2 (ACE2) complex[64] revealed key mutations on the RBD, such as F486 and N501, which form contacts with ACE2.[65] These residues are found in the pangolin coronavirus.[65]
Pangolins are protected under Chinese law, but their poaching and trading for use in traditional Chinese medicine remains common in the black market.[66][67] Deforestation, wildlife farming and trade in unsanitary conditions increases the risk of new zoonotic diseases, biodiversity experts have warned.[68][69][70]
It is unlikely that SARS-CoV-2 was genetically engineered. According to computational simulations on protein folding, the RBD of the spike protein of SARS-CoV-2 should have unremarkable binding affinity. In actuality, however, it has very efficient binding to the human ACE2 receptor. To expose the RBD for fusion, furin proteases must first cleave the S protein. Furin proteases are abundant in the respiratory tract and lung epithelial cells. Additionally, the backbone of the virus does not resemble any previously described in scientific literature used for genetic modification. The possibility that the virus could have gained the necessary adaptations through cell culture in a laboratory setting is challenged by scientists who assert that "the generation of the predicted O-linked glycans... suggest[s] the involvement of an immune system."[71][9]
Phylogenetics and taxonomy
SARS-CoV-2 belongs to the broad family of viruses known as coronaviruses.[72] It is a positive-sense single-stranded RNA (+ssRNA) virus, with a single linear RNA segment. Other coronaviruses are capable of causing illnesses ranging from the common cold to more severe diseases such as Middle East respiratory syndrome (MERS, fatality rate ~34%). It is the seventh known coronavirus to infect people, after 229E, NL63, OC43, HKU1, MERS-CoV, and the original SARS-CoV.[73]
Like the SARS-related coronavirus strain implicated in the 2003 SARS outbreak, SARS-CoV-2 is a member of the subgenus Sarbecovirus (beta-CoV lineage B).[74][75] Its RNA sequence is approximately 30,000 bases in length.[2] SARS-CoV-2 is unique among known betacoronaviruses in its incorporation of a polybasic cleavage site, a characteristic known to increase pathogenicity and transmissibility in other viruses.[9][76][77]
With a sufficient number of sequenced genomes, it is possible to reconstruct a phylogenetic tree of the mutation history of a family of viruses. By 12 January 2020, five genomes of SARS-CoV-2 had been isolated from Wuhan and reported by the Chinese Center for Disease Control and Prevention (CCDC) and other institutions;[2][78] the number of genomes increased to 42 by 30 January 2020.[79] A phylogenetic analysis of those samples showed they were "highly related with at most seven mutations relative to a common ancestor", implying that the first human infection occurred in November or December 2019.[79] Sablon:As of 4,690 SARS-CoV-2 genomes sampled on six continents were publicly available.[80]
On 11 February 2020, the International Committee on Taxonomy of Viruses announced that according to existing rules that compute hierarchical relationships among coronaviruses on the basis of five conserved sequences of nucleic acids, the differences between what was then called 2019-nCoV and the virus strain from the 2003 SARS outbreak were insufficient to make them separate viral species. Therefore, they identified 2019-nCoV as a strain of Severe acute respiratory syndrome-related coronavirus.[3]
Structural biology

Each SARS-CoV-2 virion is 50–200 nanometres in diameter.[44] Like other coronaviruses, SARS-CoV-2 has four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope.[81] The spike protein, which has been imaged at the atomic level using cryogenic electron microscopy,[82][83] is the protein responsible for allowing the virus to attach to and fuse with the membrane of a host cell;[81] specifically, its S1 subunit catalyzes attachment, the S2 subunit fusion.[84]
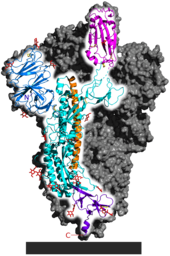
Protein modeling experiments on the spike protein of the virus soon suggested that SARS-CoV-2 has sufficient affinity to the receptor angiotensin converting enzyme 2 (ACE2) on human cells to use them as a mechanism of cell entry.[85] By 22 January 2020, a group in China working with the full virus genome and a group in the United States using reverse genetics methods independently and experimentally demonstrated that ACE2 could act as the receptor for SARS-CoV-2.[6][86][15][87] Studies have shown that SARS-CoV-2 has a higher affinity to human ACE2 than the original SARS virus strain.[82][88] SARS-CoV-2 may also use basigin to assist in cell entry.[89]
Initial spike protein priming by transmembrane protease, serine 2 (TMPRSS2) is essential for entry of SARS-CoV-2.[16] After a SARS-CoV-2 virion attaches to a target cell, the cell's protease TMPRSS2 cuts open the spike protein of the virus, exposing a fusion peptide in the S2 subunit, and the host receptor ACE2.[84] After fusion, an endosome forms around the virion, separating it from the rest of the host cell. The virion escapes when the pH of the endosome drops or when cathepsin, a host cysteine protease, cleaves it.[84] The virion then releases RNA into the cell and forces the cell to produce and disseminate copies of the virus, which infect more cells.[90]
SARS-CoV-2 produces at least three virulence factors that promote shedding of new virions from host cells and inhibit immune response.[81] Whether they include downregulation of ACE2, as seen in similar coronaviruses, remains under investigation (as of May 2020).[57]
{{Több kép}}: nincs elég kép
Epidemiology

Based on the low variability exhibited among known SARS-CoV-2 genomic sequences, the strain is thought to have been detected by health authorities within weeks of its emergence among the human population in late 2019.[12][91] The earliest case of infection currently known is dated back to 17 November 2019 or possibly 1 December 2019.[92] The virus subsequently spread to all provinces of China and to more than 150 other countries in Asia, Europe, North America, South America, Africa, and Oceania.[93] Human-to-human transmission of the virus has been confirmed in all these regions.[94] On 30 January 2020, SARS-CoV-2 was designated a Public Health Emergency of International Concern by the WHO,[95][96] and on 11 March 2020 the WHO declared it a pandemic.[97][98]
The basic reproduction number () of the virus has been estimated to be between 1.4 and 3.9.[99][100] This means each infection from the virus is expected to result in 1.4 to 3.9 new infections when no members of the community are immune and no preventive measures are taken. The reproduction number may be higher in densely populated conditions such as those found on cruise ships.[101] Many forms of preventive efforts may be employed in specific circumstances in order to reduce the propagation of the virus.
There have been about 82,000 confirmed cases of infection in mainland China.[93] While the proportion of infections that result in confirmed cases or progress to diagnosable disease remains unclear,[102] one mathematical model estimated that 75,815 people were infected on 25 January 2020 in Wuhan alone, at a time when the number of confirmed cases worldwide was only 2,015.[103] Before 24 February 2020, over 95% of all deaths from COVID-19 worldwide had occurred in Hubei province, where Wuhan is located.[104][105] As of Sablon:Cases in the COVID-19 pandemic, the percentage had decreased to Sablon:Percentage.Sablon:Cases in the COVID-19 pandemic
As of Sablon:Cases in the COVID-19 pandemic, there have been Sablon:Cases in the COVID-19 pandemic total confirmed cases of SARS-CoV-2 infection in the ongoing pandemic.Sablon:Cases in the COVID-19 pandemic The total number of deaths attributed to the virus is Sablon:Cases in the COVID-19 pandemic.Sablon:Cases in the COVID-19 pandemic Many recoveries from confirmed infections go unreported, but at least Sablon:Cases in the COVID-19 pandemic people have recovered from confirmed infections.Sablon:Cases in the COVID-19 pandemic
See also
- Decoding COVID-19 – 2020 PBS film documentary about the 2019–2020 COVID-19 pandemic
References
- ↑ (1971. május 6.) „Expression of animal virus genomes.”. Bacteriological Reviews 35 (3), 235–241. o. DOI:10.1128/MMBR.35.3.235-241.1971. PMID 4329869.
- ↑ a b c CoV2020. GISAID EpifluDB . [2020. január 12-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. január 12.)
- ↑ a b (2020. március 17.) „Correspondence: Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1”. The New England Journal of Medicine 382 (16), 1564–1567. o. DOI:10.1056/NEJMc2004973. PMID 32182409.
- ↑ New coronavirus stable for hours on surfaces (angol nyelven). National Institutes of Health (NIH) . NIH.gov, 2020. március 17. [2020. március 23-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. május 4.)
- ↑ a b c d e (2020. február 1.) „A pneumonia outbreak associated with a new coronavirus of probable bat origin”. Nature 579 (7798), 270–273. o. DOI:10.1038/s41586-020-2012-7. PMID 32015507.
- ↑ (2020. február 1.) „Another Decade, Another Coronavirus”. The New England Journal of Medicine 382 (8), 760–762. o. DOI:10.1056/NEJMe2001126. PMID 31978944.
- ↑ a b (2020. április 1.) „The 2019-new coronavirus epidemic: Evidence for virus evolution”. Journal of Medical Virology 92 (4), 455–459. o. DOI:10.1002/jmv.25688. PMID 31994738.
- ↑ a b c d e (2020. március 17.) „Correspondence: The proximal origin of SARS-CoV-2”. Nature Medicine 26 (4), 450–452. o. DOI:10.1038/s41591-020-0820-9. PMID 32284615.
- ↑ (2020. február 11.) „Novel Coronavirus (2019-nCoV): situation report, 22”, Kiadó: World Health Organization.
- ↑ „Coronavirus: From bats to pangolins, how do viruses reach us?”, Deutsche Welle, 2020. február 7.. [2020. június 4-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 13.)
- ↑ a b c d (2020. január 1.) „Wuhan seafood market may not be source of novel virus spreading globally”. Science. DOI:10.1126/science.abb0611.
- ↑ Q&A on coronaviruses (COVID-19). World Health Organization (WHO) , 2020. február 11. [2020. január 20-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. február 24.)
- ↑ a b How COVID-19 Spreads. U.S. Centers for Disease Control and Prevention (CDC) , 2020. január 27. [2020. január 28-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. január 29.)
- ↑ a b (2020. február 1.) „Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses”. Nature Microbiology 5 (4), 562–569. o. DOI:10.1038/s41564-020-0688-y. PMID 32094589.
- ↑ a b (2020. április 16.) „SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor”. Cell 181 (2), 271–280.e8. o. DOI:10.1016/j.cell.2020.02.052. PMID 32142651.
- ↑ Wu, Katherine J.. „There are more viruses than stars in the universe. Why do only some infect us? - More than a quadrillion quadrillion individual viruses exist on Earth, but most are not poised to hop into humans. Can we find the ones that are?”, National Geographic Society, 2020. április 15.. [2020. április 23-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. május 18.)
- ↑ (2020. február 1.) „A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster”. The Lancet 395 (10223), 514–523. o. DOI:10.1016/S0140-6736(20)30154-9. PMID 31986261.
- ↑ (2020. március 1.) „The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future”. Microbes and Infection 22 (2), 80–85. o. DOI:10.1016/j.micinf.2020.02.002. PMID 32087334. (Hozzáférés: 2020. április 19.)
- ↑ Kessler, Glenn. „Trump's false claim that the WHO said the coronavirus was 'not communicable'”, The Washington Post, 2020. április 17.. [2020. április 17-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. április 17.)
- ↑ Kuo, Lily. „China confirms human-to-human transmission of coronavirus”, The Guardian, 2020. január 21.. [2020. március 22-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. április 18.)
- ↑ „How does coronavirus spread?”, NBC News, 2020. január 25.. [2020. január 28-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 13.)
- ↑ (2020. május 1.) „Visualizing Speech-Generated Oral Fluid Droplets with Laser Light Scattering”. The New England Journal of Medicine 382 (21), 2061–2063. o. DOI:10.1056/NEJMc2007800. PMID 32294341.
- ↑ (2020. június 1.) „The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission”. Proceedings of the National Academy of Sciences of the United States of America 117 (22), 11875–11877. o. DOI:10.1073/pnas.2006874117. PMID 32404416.
- ↑ Apoorva Mandavilli, Apoorva]]. „239 Experts With One Big Claim: The Coronavirus Is Airborne - The W.H.O. has resisted mounting evidence that viral particles floating indoors are infectious, some scientists say. The agency maintains the research is still inconclusive.”, The New York Times, 2020. július 4. (Hozzáférés: 2020. július 5.)
- ↑ Getting your workplace ready for COVID-19. World Health Organization , 2020. február 27. [2020. március 2-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. március 3.)
- ↑ Why the Coronavirus Has Been So Successful. The Atlantic , 2020. március 20. [2020. március 20-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. március 20.)
- ↑ Why soap is preferable to bleach in the fight against coronavirus. National Geographic , 2020. március 18. [2020. április 2-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. április 2.)
- ↑ (2020. március 1.) „First Case of 2019 Novel Coronavirus in the United States”. The New England Journal of Medicine 382 (10), 929–936. o. DOI:10.1056/NEJMoa2001191. PMID 32004427.
- ↑ (2020. május 7.) „Clinical Characteristics and Results of Semen Tests Among Men With Coronavirus Disease 2019”. JAMA Network Open 3 (5), e208292. o. DOI:10.1001/jamanetworkopen.2020.8292. PMID 32379329.
- ↑ (2020. április 1.) „Virological assessment of hospitalized patients with COVID-2019”. Nature 581 (7809), 465–469. o. DOI:10.1038/s41586-020-2196-x. PMID 32235945.
- ↑ (2020. február 1.) „Study claiming new coronavirus can be transmitted by people without symptoms was flawed”. Science. DOI:10.1126/science.abb1524.
- ↑ (2020. március 1.) „Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study”. The Lancet Infectious Diseases 20 (5), 565–574. o. DOI:10.1016/S1473-3099(20)30196-1. PMID 32213337. (Hozzáférés: 2020. április 21.)
- ↑ (2020. február 1.) „Novel Coronavirus (2019-nCoV): situation report, 12”, Kiadó: World Health Organization.
- ↑ (2020. március 16.) „Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2)”. Science 368 (6490), 489–493. o. DOI:10.1126/science.abb3221. PMID 32179701.
- ↑ Daily Telegraph, Thursday 28 May 2020, page 2 column 1, which refers to the medical journal Thorax; Thorax May 2020 article COVID-19: in the footsteps of Ernest Shackleton Archiválva 2020. május 30-i dátummal a Wayback Machine-ben.
- ↑ (2020. április 15.) „Temporal dynamics in viral shedding and transmissibility of COVID-19”. Nature Medicine 26 (5), 672–675. o. DOI:10.1038/s41591-020-0869-5. PMID 32296168. (Hozzáférés: 2020. április 21.)
- ↑ Questions and Answers on the COVID-19: OIE - World Organisation for Animal Health. www.oie.int . [2020. március 31-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. április 16.)
- ↑ Goldstein, Joseph. „Bronx Zoo Tiger Is Sick with the Coronavirus”, The New York Times, 2020. április 6.. [2020. április 9-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. április 10.)
- ↑ USDA Statement on the Confirmation of COVID-19 in a Tiger in New York. United States Department of Agriculture , 2020. április 5. [2020. április 15-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. április 16.)
- ↑ If You Have Animals—Coronavirus Disease 2019 (COVID-19) (amerikai angol nyelven). Centers for Disease Control and Prevention (CDC) , 2020. április 13. [2020. április 1-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. április 16.)
- ↑ We're still not sure where the Wuhan coronavirus really came from. Popular Science , 2020. január 28. [2020. január 30-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. január 30.)
- ↑ (2020. február 15.) „Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China”. The Lancet 395 (10223), 497–506. o. DOI:10.1016/S0140-6736(20)30183-5. PMID 31986264. (Hozzáférés: 2020. március 26.)
- ↑ a b (2020. február 15.) „Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study”. The Lancet 395 (10223), 507–513. o. DOI:10.1016/S0140-6736(20)30211-7. PMID 32007143. (Hozzáférés: 2020. március 9.)
- ↑ a b (2020. február 26.) „Mystery deepens over animal source of coronavirus”. Nature 579 (7797), 18–19. o. DOI:10.1038/d41586-020-00548-w. PMID 32127703.
- ↑ (2020. február 21.) „Decoding evolution and transmissions of novel pneumonia coronavirus using the whole genomic data”. DOI:10.12074/202002.00033. (Hozzáférés: 2020. február 25.)
- ↑ (2020. április 8.) „Phylogenetic network analysis of SARS-CoV-2 genomes” (angol nyelven). PNAS 117 (17), 9241–9243. o. DOI:10.1073/pnas.2004999117. PMID 32269081. (Hozzáférés: 2020. április 17.)
- ↑ COVID-19: genetic network analysis provides 'snapshot' of pandemic origins (angol nyelven). Cambridge University , 2020. április 9. [2020. április 16-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. április 17.)
- ↑ (2020. február 15.) „Bat SARS-like coronavirus isolate bat-SL-CoVZC45, complete genome”. (Hozzáférés: 2020. február 15.)
- ↑ (2020. február 15.) „Bat SARS-like coronavirus isolate bat-SL-CoVZXC21, complete genome”. (Hozzáférés: 2020. február 15.)
- ↑ Bat coronavirus isolate RaTG13, complete genome. National Center for Biotechnology Information (NCBI) , 2020. február 10. [2020. május 15-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. március 5.)
- ↑ (2020. február 24.) „Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19)”, Kiadó: World Health Organization (WHO). (Hozzáférés: 2020. március 5.)
- ↑ (2020. február 1.) „Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding”. The Lancet 395 (10224), 565–574. o. DOI:10.1016/S0140-6736(20)30251-8. PMID 32007145.
- ↑ (2020. március 12.) „The SARS-CoV-2 outbreak: what we know” (english nyelven). International Journal of Infectious Diseases 94, 44–48. o. DOI:10.1016/j.ijid.2020.03.004. ISSN 1201-9712. PMID 32171952. (Hozzáférés: 2020. április 16.)
- ↑ (2020. április 1.) „Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event”. Infection, Genetics and Evolution 79, 104212. o. DOI:10.1016/j.meegid.2020.104212. PMID 32004758. (Hozzáférés: 2020. április 9.)
- ↑ a b (2020. március 19.) „Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak”. Current Biology 30 (7), 1346–1351.e2. o. DOI:10.1016/j.cub.2020.03.022. PMID 32197085.
- ↑ a b BMJ Best Practice: Coronavirus Disease 2019 (COVID-19) (angol nyelven). BMJ , 2020. május 22. [2020. június 13-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. május 25.)
- ↑ A Virologist Explains Why It Is Unlikely COVID-19 Escaped From A Lab. Forbes, 2020. április 17. [2020. április 21-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. április 22.)
- ↑ (2019. október 1.) „Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica)”. Viruses 11 (11), 979. o. DOI:10.3390/v11110979. PMID 31652964.
- ↑ (2020. február 7.) „Did pangolins spread the China coronavirus to people?”. Nature. DOI:10.1038/d41586-020-00364-2. (Hozzáférés: 2020. február 12.)
- ↑ (2020. február 1.) „Isolation and Characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins”. bioRxiv. DOI:10.1101/2020.02.17.951335. (Hozzáférés: 2020. május 5.)
- ↑ (2020. február 1.) „Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019”. bioRxiv. DOI:10.1101/2020.02.07.939207. PMID 32511310. (Hozzáférés: 2020. május 5.)
- ↑ Timmer, John: SARS-CoV-2 looks like a hybrid of viruses from two different species (amerikai angol nyelven). Ars Technica , 2020. június 1. [2020. június 5-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. június 6.)
- ↑ (2020. március 27.) „Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2”. Science 367 (6485), 1444–1448. o. DOI:10.1126/science.abb2762. ISSN 1095-9203. PMID 32132184.
- ↑ a b Ho, Mitchell (2020. április 30.). „Perspectives on the development of neutralizing antibodies against SARS-CoV-2” (angol nyelven). Antibody Therapeutics 3 (2), 109–114. o. DOI:10.1093/abt/tbaa009. PMID 32566896. (Hozzáférés: 2020. június 14.)
- ↑ „Pangolins: 13 facts about the world's most hunted animal”, The Telegraph, 2015. január 1.. [2019. december 24-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 9.)
- ↑ „China's Ban on Wildlife Trade a Big Step, but Has Loopholes, Conservationists Say”, The New York Times, 2020. február 27.. [2020. március 13-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 23.)
- ↑ Carrington, Damian. „Halt destruction of nature or suffer even worse pandemics, say world's top scientists”, The Guardian, 2020. április 27.. [2020. május 15-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. május 31.) (brit angol nyelvű)
- ↑ Pontes, Nadia: How deforestation can lead to more infectious diseases (brit angol nyelven). DW.COM , 2020. április 29. [2020. május 5-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. május 31.)
- ↑ (2007. október 1.) „Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection”. Clinical Microbiology Reviews 20 (4), 660–694. o. DOI:10.1128/CMR.00023-07. ISSN 0893-8512. PMID 17934078.
- ↑ „The COVID-19 coronavirus epidemic has a natural origin, scientists say—Scripps Research's analysis of public genome sequence data from SARS‑CoV‑2 and related viruses found no evidence that the virus was made in a laboratory or otherwise engineered”, EurekAlert!, Scripps Research Institute, 2020. március 17.. [2020. április 3-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. április 15.)
- ↑ (2020. január 24.) „What you need to know about the Wuhan coronavirus”. Nature. DOI:10.1038/d41586-020-00209-y.
- ↑ (2020. február 1.) „A Novel Coronavirus from Patients with Pneumonia in China, 2019”. The New England Journal of Medicine 382 (8), 727–733. o. DOI:10.1056/NEJMoa2001017. PMID 31978945.
- ↑ Phylogeny of SARS-like betacoronaviruses. nextstrain . [2020. január 20-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. január 18.)
- ↑ (2019. február 1.) „Global Epidemiology of Bat Coronaviruses”. Viruses 11 (2), 174. o. DOI:10.3390/v11020174. PMID 30791586.
- ↑ (2020. március 9.) „Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein”. Cell 181 (2), 281–292.e6. o. DOI:10.1016/j.cell.2020.02.058. PMID 32155444.
- ↑ Initial genome release of novel coronavirus. Virological , 2020. január 11. [2020. január 12-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. január 12.)
- ↑ a b Genomic analysis of nCoV spread: Situation report 2020-01-30. nextstrain.org . [2020. március 15-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. március 18.)
- ↑ Genomic epidemiology of novel coronavirus - Global subsampling. Nextstrain . [2020. április 20-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. május 7.)
- ↑ a b c (2020. február 1.) „Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods”. Acta Pharmaceutica Sinica B 10 (5), 766–788. o. DOI:10.1016/j.apsb.2020.02.008. PMID 32292689.
- ↑ a b (2020. február 1.) „Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation”. Science 367 (6483), 1260–1263. o. DOI:10.1126/science.abb2507. PMID 32075877.
- ↑ Scientists Create Atomic-Level Image of the New Coronavirus's Potential Achilles Heel. Gizmodo , 2020. február 19. [2020. március 8-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. március 13.)
- ↑ (2020. március 1.) „Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission”. Science China Life Sciences 63 (3), 457–460. o. DOI:10.1007/s11427-020-1637-5. PMID 32009228.
- ↑ (2020. január 1.) „Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV”. bioRxiv. DOI:10.1101/2020.01.22.915660. PMID 32511294. (Hozzáférés: 2020. május 5.)
- ↑ „Genomic Characterization of the 2019 Novel Coronavirus”. The New England Journal of Medicine. (Hozzáférés: 2020. február 9.)
- ↑ Novel coronavirus structure reveals targets for vaccines and treatments. National Institutes of Health (NIH) , 2020. március 2. [2020. április 1-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. április 3.)
- ↑ (2020. március 14.) „SARS-CoV-2 invades host cells via a novel route: CD147-spike protein”. bioRxiv. DOI:10.1101/2020.03.14.988345. (Hozzáférés: 2020. május 5.)
- ↑ Anatomy of a Killer: Understanding SARS-CoV-2 and the drugs that might lessen its power (2020. március 12.)
- ↑ What We Know Today about Coronavirus SARS-CoV-2 and Where Do We Go from Here. Genetic Engineering and Biotechnology News , 2020. február 19. [2020. március 14-i dátummal az eredetiből archiválva]. (Hozzáférés: 2020. március 13.)
- ↑ „Coronavirus: China's first confirmed Covid-19 case traced back to November 17”, 2020. március 13.. [2020. március 13-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 16.)
- ↑ a b Forráshivatkozás-hiba: Érvénytelen
<ref>címke; nincs megadva szöveg a(z)JHU_tickernevű lábjegyzeteknek - ↑ (2020. március 29.) „Coronavirus disease 2019 (COVID-19) Situation Report – 69”, Kiadó: World Health Organization.
- ↑ „W.H.O. Declares Global Emergency as Wuhan Coronavirus Spreads”, 2020. január 30.. [2020. január 30-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. január 30.)
- ↑ (30 January 2020). "Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV)". Sajtóközlemény.
- ↑ (11 March 2020). "WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020". Sajtóközlemény.
- ↑ „Coronavirus Declared Pandemic by World Health Organization”, 2020. március 11.. [2020. március 11-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 12.)
- ↑ (2020. január 1.) „Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia”. The New England Journal of Medicine 382 (13), 1199–1207. o. DOI:10.1056/NEJMoa2001316. PMID 31995857.
- ↑ (2020. január 1.) „Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020”. Eurosurveillance 25 (4). DOI:10.2807/1560-7917.ES.2020.25.4.2000058. PMID 32019669.
- ↑ (2020. február 1.) „COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures”. Journal of Travel Medicine 27 (3). DOI:10.1093/jtm/taaa030. PMID 32109273.
- ↑ „Limited data on coronavirus may be skewing assumptions about severity”, STAT, 2020. január 30.. [2020. február 1-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 13.)
- ↑ (2020. február 1.) „Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study”. The Lancet 395 (10225), 689–697. o. DOI:10.1016/S0140-6736(20)30260-9. PMID 32014114.
- ↑ „Coronavirus deaths leap in China as countries struggle to evacuate citizens”, The Guardian, 2020. január 30.. [2020. február 6-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 10.)
- ↑ „Coronavirus: China to repay Africa in safeguarding public health”, The Sun, 2020. február 25.. [2020. március 9-i dátummal az eredetiből archiválva] (Hozzáférés: 2020. március 10.)
Forráshivatkozás-hiba: a <references> címkében definiált „BBC-Named” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „CDC-nCoV” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „CDCAbout” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „COVIDname” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „EconomistSinophobia” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „HillWHOname” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „HuangNPR” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „NBC-GOP” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „NYT-SpikyBlob” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „NYT6” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „QZ-Why” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „TodayNameMixup” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „USnews17March2020” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „WHO-SR10” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „WHO21Jan2020” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „WHOnamingguidelines” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Forráshivatkozás-hiba: a <references> címkében definiált „Zoonotic” nevű <ref> címke nem szerepel a szöveg korábbi részében.
Further reading
- (2020. március 31.) „SARS-CoV-2 (COVID-19) by the numbers”. eLife 9. DOI:10.7554/eLife.57309. PMID 32228860.
- Brüssow, Harald (2020. március 1.). „The Novel Coronavirus – A Snapshot of Current Knowledge”. Microbial Biotechnology 2020 (3), 607–612. o. DOI:10.1111/1751-7915.13557. PMID 32144890.
- (2020. január 1.) „Features, Evaluation and Treatment Coronavirus (COVID-19)”. StatPearls. PMID 32150360. (Hozzáférés: 2020. április 4.)
- (2020. február 1.) „The Novel Coronavirus: A Bird's Eye View”. The International Journal of Occupational and Environmental Medicine 11 (2), 65–71. o. DOI:10.15171/ijoem.2020.1921. PMID 32020915.
- (2020. március 2.) „Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases”, Kiadó: World Health Organization.
External links
- Coronavirus Disease 2019 (COVID-19). Centers for Disease Control and Prevention (CDC) , 2020. február 11.
- Coronavirus disease (COVID-19) Pandemic. World Health Organization (WHO)
- SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) Sequences. National Center for Biotechnology Information (NCBI)
- COVID-19 Resource Centre. The Lancet
- Coronavirus (Covid-19). The New England Journal of Medicine
- Covid-19: Novel Coronavirus Outbreak. Wiley
- SARS-CoV-2. Virus Pathogen Database and Analysis Resource
- SARS-CoV-2 related protein structures. Protein Data Bank
Sablon:Medical resources Sablon:COVID-19 pandemic Sablon:Human coronaviruses Sablon:Viral systemic diseases Sablon:Zoonotic viral diseases Sablon:Subject bar

