„Mikro-RNS” változatai közötti eltérés
Új oldal, tartalma: „{{építés alatt}} bélyegkép|400px|Pre-miRNS a Pri-miRNS helyett a mechanizmus első pontjában. A mikro-RNS hatása [[mRNS-re]] bélyegkép|400px|miRNS-hurkok, az érett miRNS-ek pirossal A '''mikro-RNS''' ('''miRNS''') kis, egyszálú nem kódoló RNS 21–23 nukleotiddal.<ref name="Metazoan MicroRNAs">{{cite journal|author=Bartel DP|title=Metazoan MicroRNAs|journal=Cell|volume=173|issue=1|…” Címkék: tataroz vagy építés alatt sablon kihelyezve Hullámos ő – kalapos ű Egyértelműsítő hivatkozások |
(Nincs különbség)
|
A lap 2024. február 22., 17:34-kori változata
| Ez a szócikk/szakasz most épül, még dolgoznak az első verzión! |
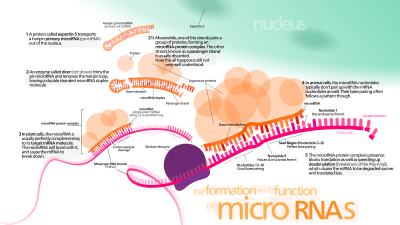

A mikro-RNS (miRNS) kis, egyszálú nem kódoló RNS 21–23 nukleotiddal.[1] Növényekben, állatokban és egyes vírusokban találhatók, és az RNS-csendesítésben és a poszttranszlációs génexpresszió-szabályzásban fontosak.[2][3] A miRNS-ek a megfelelő mRNS-ek kopmlementer szakaszaihoz kötnek,[4] majd ezeket az alábbi folyamatok egyikével csendesítik:[1][5]
- Felbontás két részre,
- Destabilizáció a poli(A)-farok rövidítése vagy
- fehérjékké való transzláció csökkentése.
Állati sejtekben a miRNS-ek elsősorban az mRNS destabilizációjával hatnak.[5][6]
A miRNS-ek a kis interferáló RNS-ekhez (siRNS) hasonlítanak az RNS-interferencia (RNSi) útjában, azonban a miRNS-ek önmagukra visszahajló rövid hajtűket alkotó RNS-transzkriptum-részekből származnak, míg a siRNS-ek hosszabb kétszálú RNS-részekből.[2] A humán genom több mint 1900 miRNS-t kódolhat,[7][8] Azonban csak mintegy 500 humán miRNS jelent bona fide miRNS-t a kézzel kezelt MirGeneDB miRNS-gén-adatbázisban.[9]
Számos emlőssejttípusban sok miRNS található:[10][11] ezek a humán és más emlősgének mintegy 60%-át célozzák.[12][13] Sok miRNS evolúciósan állandósult, vagyis fontos biológiai funkcióik lehetnek.[14][1] Például 90 miRNS-család legalább az emlősök és a halak közös őséig visszamenőleg állandósult, és ezeknek fontos funkcióik vannak, melyeket ezek egy vagy több tagjának eltávolításával végzett egérkísérletek mutattak ki.[1]
Történet
Az első miRNS-t 1993-ban fedezték fel.[15] Azonban csak 2000-ben ismerték el önálló biológiaiszabályzó-osztályként.[16][17][18][19][20] a miRNS-kutatás számos eltérő miRNS-csoportot mutatott ki eltérő sejttípusokban és szövetekben,[11][21] valamint számos szerepet a miRNS-eknek a növényi és állati fejlődésben és sok más biológiai folyamatban.[22][23][24][25][26][27][28] A nem megfelelő miRNS-expresszió számos betegségben megjelenik. Vizsgálnak miRNS-alapú terápiákat.[29][30][31][32]
Az első miRNS-t 1993-ban fedezte fel Victor Ambros, Lee és Feinbaum. Azonban további vizsgálatához Gary Ruvkun, Wightman és Ha munkája kellett.[15][33] E csoportok egymás után közöltek tanulmányokat a lin-4 génről, mely a Caenorhabditis elegans-lárva fejlődését irányítja a lin-14 gén repressziójával. Miután Lee et al. izolálták a lin-4 miRNS-t, észrevették, hogy nem fehérjekódoló mRNS-t kódol, hanem rövid nem kódoló RNS-t, melyek egyike a lin-14-mRNS 3'-transzlálatlanrégiójának több szakaszával részben komplementer.[15] Feltételezésük szerint a lin-4 a lin-14 mRNS LIN-14 fehérjévé való transzlációját gátolta, és úgy gondolták, hogy a lin-4 csak gyűrűsférgekre jellemző.
2000-ben fedezték fel a második kis RNS-t, a let-7-RNS-t, mely a lin-41-et represszálja a C. elegans későbbi fejlődési átmenetéhez.[16] Ezt sok fajban állandósultnak találták, ahhoz a feltételezéshez vezetve, hogy a let-7-RNS és más „kis temporális RNS-ek” számos állat – beleértve az embert – fejlődésének időzítését szabályozhatják.[17]
2001-ben a lin-4 és let-7 RNS-ekről kiderítették, hogy egy, a C. elegans, a Drosophila és az ember sejtjeiben jelenlévő nagy kis-RNS-osztály tagja.[18][19][20] Ennek sok RNS-e hasonlított a lin-4 és let-7-RNS-ekre, eltekintve a fejlődésidőzítés szabályzásának nem megfelelő expressziós mintával. Ez alapján feltételezték, hogy a legtöbb más szabályzó utakban vesz részt. Ekkor kezdték el a kis szabályzó RNS-eket mikro-RNS-nek nevezni.[18][19][20]
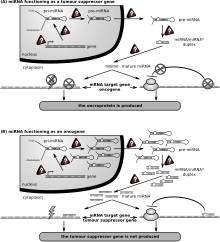
Az első miRNS-deregulációval összefüggő humán betegség a B-sejtes krónikus limfocitás leukémia volt. E betegségben a miRNS-ek egyszerre működnek tumorszupresszorként és onkogénként.[34]
Nevezéktan
A szabványos nevezéktanban a kísérletileg igazolt miRNS-ek nevüket közlés előtt kapják.[35][36] A „miR” prefixumot kötőjel, majd szám követi, ez utóbbi gyakran a megnevezés sorrenjét jelenti. Például a miR-124 korábban kapta nevét, és korábban fedezték fel a miR-456-nál. A nagybetűs „miR-” érett miRNS-t jelent, a nagybetű nélküli „mir-” a pre- és pri-miRNS-re.[37] A miRNS-kódoló gének ugyane 3 betűs prefixumot kapják génnevezéktanuknak megfelelően – példák miRNS-gén-nevekre a mir-1 C. elegans és Drosophila esetén, Mir1 Rattus norvegicus esetén és MIR25 emberben.
Az egy-két nukleotid kivételével teljesen eltérő szekvenciák további kisbetűt kapnak. Például a miR-124a a miR-124b közeli rokona. Például:
- hsa-miR-181a: 5’-aacauucaACgcugucggugAgu-3’
- hsa-miR-181b: 5’-aacauucaUUgcugucggugGgu-3’
A pre-, pri-miRNS-ek és az azonos érett miRNS-hez vezető különböző helyű gének további kötőjelet és számot kapnak. Például a hsa-mir-194-1 és hsa-mir-194-2 azonos érett miRNS-t (hsa-miR-194) hoz létre, de eltérő genombeli helyről származnak.
A faj hárombetűs prefixummal van jelölve – például a hsa-miR-124 humán, az oar-miR-123 bárány-miRNS. További prefixumok például a vírusokat jelölő v vagy a Drosophilát jelölő d.
Ha két érett miRNS azonos pre-miRNS ellentétes karjairól származik és nagyjából ugyanannyi van belőlük, -3p vagy -5p utótagot kapnak (korábban s (szenz) vagy as (antiszenz) utótaggal is jelölték). Azonban a hajtű egyik karjáról származó miRNS általában sokkal nagyobb mennyiségben van jelen, mint a másikról származó,[2] ez esetben a nevet követő csillag a kisebb mennyiségben a másik karról keletkező miRNS-t jelenti. Például a miR-124 és a miR-124* azonos pre-miRNS hajtűvel rendelkeznek, de sokkal több a miR-124.
Célok
A növényi miRNS-ek közel tökéletesen párosodnak mRNS céljaikkal, a génrepressziót a célpont lebontásával elérve.[22][38] Ezzel szemben az állati miRNS-ek cél-mRNS-üket csupán 6–8 nukleotiddal (magrégió) ismerik fel a kezdetükön,[12][39][40] ami kevés a cél-mRNS-bontás elindításához.[4] A kombinációszabályzás fontos az állatok miRNS-szabályzásában.[4][41] Egy adott miRNS több száz mRNS-célponttal rendelkezhet, és egy célpontot több miRNS szabályozhatja.[13][42]
Adott miRNS által célzott egyedi mRNS-ek átlagos száma becsléstől függően eltér,[43] de több megközelítés szerint az emlős-miRNS-ek számos egyedi célponttal rendelkezhetnek. Például a gerincesekben állandósult miRNS-eknek átlagosan mintegy 400 állandó célpontjuk van.[13] Ugyanígy kísérletek szerint egy miRNS több száz egyedi mRNS stabilitását csökkentheti.[44] Más kísérletek szerint egy miRNS több száz fehérje termelését represszálhatja, de gyakran ez enyhe – kevesebb mint 2-szeres.[45][46]
Szintézis

A miRNS-gének akár 40%-a más gének intronjaiban vagy akár exonjaiban lehet.[47] Ezek gyakran szenz elrendezésben vannak,[48][49] így szabályzásuk a gazdagénekével együtt történik.[47][50][51]
A DNS-templáttal nem ér véget a miRNS-érés: a humán miRNS-ek 6%-a helyspecifikus RNS-szerkesztést mutat (izomiR) a DNS által kódolttól eltérő termékek létrehozásához. Ez a genom által kódolthoz képest növeli a miRNS-hatás diverzitását.
Transzkripció
A miRNS-géneket általában az RNS-polimeráz II (Pol II) írja át.[52][53] Gyakran köt a szekvenciához közeli promotert, mely a pre-miRNS hajtűhurka. Az így létrejött átirat 5'-végén speciálisan módosult nukleotid jön létre, poli(A)-farkat kap,[52][48] és leválik. Az állati miRNS-ek kezdetben egy mintegy 80 nukleotidos RNS-törzskörben keletkeznek, mely egy néhány száz nukleotidos miRNS-prekurzor, a pri-miRNS része.[52][48] Ha egy törzskörprekurzor van a 3'-UTR-ben, az átirat pri-miRNS és mRNS is lehet.[48] Az RNS-polimeráz III (Pol III) néhány miRNS-t átír, különösen a korai Alu-szekvenciákkal rendelkező, a transzfer- és az emlős-szélesszórtismétlődés (MWIR) promotereket.[54]
Magi feldolgozás

Egy pri-miRNS 1–6 miRNS-prekurzort tartalmazhat. Ezek mintegy 70 nukleotidból állnak, és a hatékony feldolgozáshoz szükséges szekvenciák veszik körül.
A kétszálú RNS-szerkezetet (dsRNS) a pri-miRNS-ek hajtűiben a DiGeorge-szindróma, 8. fontos régió (DGCR8, gerinctelenekben Pasha) veszi körül, mely nevét a DiGeorge-szindrómával való kapcsolatáról kapta. Ez az RNS-vágó Drosha enzimmel asszociál mikroprocesszor-komplexet alkotva.[55][56] E komplexben a DGCR8 a katalitikus RNáz III-domént hajtűk pri-miRNS-ekről való leválasztására használja mintegy 11 nukleotiddal a hajtűalap előtt (egy helikális dsRNS-fordulattal a törzsön belül).[57][58] A termék kétnukleotidos többlettel rendelkezik a 3'-végen, 3'-hidroxil- és 5'-foszfátcsoportjai vannak. Gyakran ezt pre-miRNS-nek nevezik. Ismertek a pre-miRNS utáni fontos szekvenciamotívumok.[59][60][61]
Az intronokból közvetlenül a mikroprocesszor-komplex megkerülésével leválasztott pre-miRNS-ek a mirtronok.[62] Ismertek mirtronok a Drosophilában, a C. elegansban és emlősökben.[63][64]
A pre-miRNS-ek 16%-át módosíthatja a magi RNS-szerkesztés.[65][66][67] A leggyakoribb az RNS-re ható adenozin-deaminázok (ADAR) általi módosítás, melyek az adenozint inozinná (A→I) alakítják. Az RNS-szerkesztés leállíthatja a magi folyamatot (például a pri-miR-142-ben a Tudor-SN ribonukleáz általi bomláshoz vezet) és a későbbi folyamatokat, például a citoplazmatikus miRNS-feldolgozást és a célspecificitást (például a miR-376 magrégiójának változtatásával a központi idegrendszerben).[65]
Magi kivitel

A pre-miRNS-hajtűk a magból a nukleocitoplazmatikus szállító exportin-5-öt tartalmazó folyamatban távoznak. E fehérje a karioferinek családjának tagja, és felismeri a Drosha által megmaradt kétnukleotidos többletet a per-miRNS-hajtű 3'-végén. Az exportin-5-mediált transzport energiafüggő, a Ran fehérjéhez kötött guanozin-trifoszfátot használ.[68]
Citoplazmatikus feldolgozás
A citoplazmában a pre-miRNS hajtűt a Dicer RNáz III bontja.[69] Ez az endoribonukleáz a hajtű 5'- és 3'-végeivel kölcsönhat,[70] és elvágja a 3'- és 5'-karokat összekötő kapcsolatokat, mintegy 22 nukleotid hosszú nem tökéletes miRNS:miRNS* duplexet adva.[69] A teljes hajtűhossz és a körméret befolyásolják a Dicer hatékonyságát. Hogy a miRNS:miRNS* pár nem tökéletes, befolyásolja az elválást is.[69][71] Egyes G-gazdag pre-miRNS-ek G-kvadruplex szerkezetet is alkothatnak a törzskörszerkezet miatt – például a humán pre-miRNS 92b a Dicer-mediált bontásnak ellenálló G-kvadruplexet alkot a citoplazmában.[72] Bár a duplex bármely szála funkcionális miRNS-ként működhet, csak egy szál kerül az RNS-indukált csendesítőkomplexbe, ahol a miRNS és mRNS célja kölcsönhatnak.
Míg a legtöbb miRNS a sejtben van, a keringő vagy extracelluláris miRNS-ek sejten kívül is megtalálhatók, beleértve számos biológiai folyadékot és sejtkultúraközeget.[73][74]
Szintézise növényekben
A miRNS-szintézis növényekben főleg a magi feldolgozásban és kivitelben tér el az állatokétól. Nem két eltérő enzim bontja a miRNS-eket egyszer a magban és egyszer azon kívül, hanem mindkét bontást a Dicer-like1 (DL1) enzim végzi. Ez csak a növényi sejtek magjában expresszálódik, így feltehetően mindkét reakció a magban történik. A növényi miRNS:miRNS* duplexek magból való kivitele előtt a 3'-végeket az RNS-metiltranszferáz Hua-erősítő 1 (HEN1) metilálja. Ezt az exportin-5-homológ Hasty (HST) szállítja a magból a citoplazmába, ahol kettéválnak, az érett miRNS a RISC-be kerül.[75]
RNS-indukált csendesítőkomplex
Az érett miRNS aktív RNS-indukált csendesítőkomplex (RISC) része, ahol a Dicer és sok kapcsolódó fehérje van.[76] A RISC-et nevezik mikro-RNS-ribonukleoprotein-komplexnek (miRNP) is.[77] A miRNS-sel rendelkező RISC-et nevezik miRISC-nek is.
A pre-miRNS Dicer általi feldolgozása feltehetően a duplex kibontásával együtt történik. Általában egy szál van a miRISC-ben, melyet termodinamikai instabilitás és a másik szálnál gyengébb 5'-végi bázispárosodás alapján választ ki.[78][79][80] A törzskör helye is befolyásolhatja a szálválasztást.[81] The other strand, called the passenger strand due to its lower levels in the steady state, is denoted with an asterisk (*) and is normally degraded. In some cases, both strands of the duplex are viable and become functional miRNA that target different mRNA populations.[82]
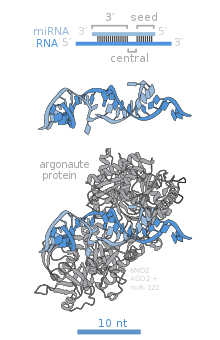
Members of the Argonaute (Ago) protein family are central to RISC function. Argonautes are needed for miRNA-induced silencing and contain two conserved RNA binding domains: a PAZ domain that can bind the single stranded 3' end of the mature miRNA and a PIWI domain that structurally resembles ribonuclease-H and functions to interact with the 5' end of the guide strand. They bind the mature miRNA and orient it for interaction with a target mRNA. Some argonautes, for example human Ago2, cleave target transcripts directly; argonautes may also recruit additional proteins to achieve translational repression.[83] The human genome encodes eight argonaute proteins divided by sequence similarities into two families: AGO (with four members present in all mammalian cells and called E1F2C/hAgo in humans), and PIWI (found in the germline and hematopoietic stem cells).[77][83]
Additional RISC components include TRBP [human immunodeficiency virus (HIV) transactivating response RNA (TAR) binding protein],[84] PACT (protein activator of the interferon-induced protein kinase), the SMN complex, fragile X mental retardation protein (FMRP), Tudor staphylococcal nuclease-domain-containing protein (Tudor-SN), the putative DNA helicase MOV10, and the RNA recognition motif containing protein TNRC6B.[68][85][86]
Mode of silencing and regulatory loops
Gene silencing may occur either via mRNA degradation or preventing mRNA from being translated. For example, miR16 contains a sequence complementary to the AU-rich element found in the 3'UTR of many unstable mRNAs, such as TNF alpha or GM-CSF.[87] It has been demonstrated that given complete complementarity between the miRNA and target mRNA sequence, Ago2 can cleave the mRNA and lead to direct mRNA degradation. In the absence of complementarity, silencing is achieved by preventing translation.[44] The relation of miRNA and its target mRNA can be based on the simple negative regulation of a target mRNA, but it seems that a common scenario is the use of a "coherent feed-forward loop", "mutual negative feedback loop" (also termed double negative loop) and "positive feedback/feed-forward loop". Some miRNAs work as buffers of random gene expression changes arising due to stochastic events in transcription, translation and protein stability. Such regulation is typically achieved by the virtue of negative feedback loops or incoherent feed-forward loop uncoupling protein output from mRNA transcription.
Turnover
Turnover of mature miRNA is needed for rapid changes in miRNA expression profiles. During miRNA maturation in the cytoplasm, uptake by the Argonaute protein is thought to stabilize the guide strand, while the opposite (* or "passenger") strand is preferentially destroyed. In what has been called a "Use it or lose it" strategy, Argonaute may preferentially retain miRNAs with many targets over miRNAs with few or no targets, leading to degradation of the non-targeting molecules.[88]
Decay of mature miRNAs in Caenorhabditis elegans is mediated by the 5'-to-3' exoribonuclease XRN2, also known as Rat1p.[89] In plants, SDN (small RNA degrading nuclease) family members degrade miRNAs in the opposite (3'-to-5') direction. Similar enzymes are encoded in animal genomes, but their roles have not been described.[88]
Several miRNA modifications affect miRNA stability. As indicated by work in the model organism Arabidopsis thaliana (thale cress), mature plant miRNAs appear to be stabilized by the addition of methyl moieties at the 3' end. The 2'-O-conjugated methyl groups block the addition of uracil (U) residues by uridyltransferase enzymes, a modification that may be associated with miRNA degradation. However, uridylation may also protect some miRNAs; the consequences of this modification are incompletely understood. Uridylation of some animal miRNAs has been reported. Both plant and animal miRNAs may be altered by addition of adenine (A) residues to the 3' end of the miRNA. An extra A added to the end of mammalian miR-122, a liver-enriched miRNA important in hepatitis C, stabilizes the molecule and plant miRNAs ending with an adenine residue have slower decay rates.[88]
Cellular functions

The function of miRNAs appears to be in gene regulation. For that purpose, a miRNA is complementary to a part of one or more messenger RNAs (mRNAs). Animal miRNAs are usually complementary to a site in the 3' UTR whereas plant miRNAs are usually complementary to coding regions of mRNAs.[91] Perfect or near perfect base pairing with the target RNA promotes cleavage of the RNA.[92] This is the primary mode of plant miRNAs.[93] In animals the match-ups are imperfect.
For partially complementary microRNAs to recognise their targets, nucleotides 2–7 of the miRNA (its 'seed region'[12][39]) must be perfectly complementary.[94] Animal miRNAs inhibit protein translation of the target mRNA[95] (this is present but less common in plants).[93] Partially complementary microRNAs can also speed up deadenylation, causing mRNAs to be degraded sooner.[96] While degradation of miRNA-targeted mRNA is well documented, whether or not translational repression is accomplished through mRNA degradation, translational inhibition, or a combination of the two is hotly debated. Recent work on miR-430 in zebrafish, as well as on bantam-miRNA and miR-9 in Drosophila cultured cells, shows that translational repression is caused by the disruption of translation initiation, independent of mRNA deadenylation.[97][98]
miRNAs occasionally also cause histone modification and DNA methylation of promoter sites, which affects the expression of target genes.[99][100]
Nine mechanisms of miRNA action are described and assembled in a unified mathematical model:[90]
- Cap-40S initiation inhibition;
- 60S Ribosomal unit joining inhibition;
- Elongation inhibition;
- Ribosome drop-off (premature termination);
- Co-translational nascent protein degradation;
- Sequestration in P-bodies;
- mRNA decay (destabilisation);
- mRNA cleavage;
- Transcriptional inhibition through microRNA-mediated chromatin reorganization followed by gene silencing.
It is often impossible to discern these mechanisms using experimental data about stationary reaction rates. Nevertheless, they are differentiated in dynamics and have different kinetic signatures.[90]
Unlike plant microRNAs, the animal microRNAs target diverse genes.[39] However, genes involved in functions common to all cells, such as gene expression, have relatively fewer microRNA target sites and seem to be under selection to avoid targeting by microRNAs.[101] There is a strong correlation between ITPR gene regulations and mir-92 and mir-19.[102]
dsRNA can also activate gene expression, a mechanism that has been termed "small RNA-induced gene activation" or RNAa. dsRNAs targeting gene promoters can induce potent transcriptional activation of associated genes. This was demonstrated in human cells using synthetic dsRNAs termed small activating RNAs (saRNAs),[103] but has also been demonstrated for endogenous microRNA.[104]
Interactions between microRNAs and complementary sequences on genes and even pseudogenes that share sequence homology are thought to be a back channel of communication regulating expression levels between paralogous genes (genes having a similar structure indicating divergence from a common ancestral gene). Given the name "competing endogenous RNAs" (ceRNAs), these microRNAs bind to "microRNA response elements" on genes and pseudogenes and may provide another explanation for the persistence of non-coding DNA.[105]
miRNAs are also found as extracellular circulating miRNAs.[106] Circulating miRNAs are released into body fluids including blood and cerebrospinal fluid and have the potential to be available as biomarkers in a number of diseases.[106][107]Some researches show that mRNA cargo of exosomes may have a role in implantation, they can savage an adhesion between trophoblast and endometrium or support the adhesion by down regulating or up regulating expression of genes involved in adhesion/invasion.[108]
Moreover, miRNA as miR-183/96/182 seems to play a key role in circadian rhythm.[109]
Evolution
miRNAs are well conserved in both plants and animals, and are thought to be a vital and evolutionarily ancient component of gene regulation.[110][111][112][113][114] While core components of the microRNA pathway are conserved between plants and animals, miRNA repertoires in the two kingdoms appear to have emerged independently with different primary modes of action.[115][116]
microRNAs are useful phylogenetic markers because of their apparently low rate of evolution.[117] microRNAs' origin as a regulatory mechanism developed from previous RNAi machinery that was initially used as a defense against exogenous genetic material such as viruses.[118] Their origin may have permitted the development of morphological innovation, and by making gene expression more specific and 'fine-tunable', permitted the genesis of complex organs[119] and perhaps, ultimately, complex life.[114] Rapid bursts of morphological innovation are generally associated with a high rate of microRNA accumulation.[117][119]
New microRNAs are created in multiple ways. Novel microRNAs can originate from the random formation of hairpins in "non-coding" sections of DNA (i.e. introns or intergene regions), but also by the duplication and modification of existing microRNAs.[120] microRNAs can also form from inverted duplications of protein-coding sequences, which allows for the creation of a foldback hairpin structure.[121] The rate of evolution (i.e. nucleotide substitution) in recently originated microRNAs is comparable to that elsewhere in the non-coding DNA, implying evolution by neutral drift; however, older microRNAs have a much lower rate of change (often less than one substitution per hundred million years),[114] suggesting that once a microRNA gains a function, it undergoes purifying selection.[120] Individual regions within an miRNA gene face different evolutionary pressures, where regions that are vital for processing and function have higher levels of conservation.[122] At this point, a microRNA is rarely lost from an animal's genome,[114] although newer microRNAs (thus presumably non-functional) are frequently lost.[120] In Arabidopsis thaliana, the net flux of miRNA genes has been predicted to be between 1.2 and 3.3 genes per million years.[123] This makes them a valuable phylogenetic marker, and they are being looked upon as a possible solution to outstanding phylogenetic problems such as the relationships of arthropods.[124] On the other hand, in multiple cases microRNAs correlate poorly with phylogeny, and it is possible that their phylogenetic concordance largely reflects a limited sampling of microRNAs.[125]
microRNAs feature in the genomes of most eukaryotic organisms, from the brown algae[126] to the animals. However, the difference in how these microRNAs function and the way they are processed suggests that microRNAs arose independently in plants and animals.[127]
Focusing on the animals, the genome of Mnemiopsis leidyi[128] appears to lack recognizable microRNAs, as well as the nuclear proteins Drosha and Pasha, which are critical to canonical microRNA biogenesis. It is the only animal thus far reported to be missing Drosha. MicroRNAs play a vital role in the regulation of gene expression in all non-ctenophore animals investigated thus far except for Trichoplax adhaerens, the only known member of the phylum Placozoa.[129]
Across all species, in excess of 5000 different miRNAs had been identified by March 2010.[130] Whilst short RNA sequences (50 – hundreds of base pairs) of a broadly comparable function occur in bacteria, bacteria lack true microRNAs.[131]
Experimental detection and manipulation
While researchers focused on miRNA expression in physiological and pathological processes, various technical variables related to microRNA isolation emerged. The stability of stored miRNA samples has been questioned.[74] microRNAs degrade much more easily than mRNAs, partly due to their length, but also because of ubiquitously present RNases. This makes it necessary to cool samples on ice and use RNase-free equipment.[132]
microRNA expression can be quantified in a two-step polymerase chain reaction process of modified RT-PCR followed by quantitative PCR. Variations of this method achieve absolute or relative quantification.[133] miRNAs can also be hybridized to microarrays, slides or chips with probes to hundreds or thousands of miRNA targets, so that relative levels of miRNAs can be determined in different samples.[134] microRNAs can be both discovered and profiled by high-throughput sequencing methods (microRNA sequencing).[135] The activity of an miRNA can be experimentally inhibited using a locked nucleic acid (LNA) oligo, a Morpholino oligo[136][137] or a 2'-O-methyl RNA oligo.[138] A specific miRNA can be silenced by a complementary antagomir. microRNA maturation can be inhibited at several points by steric-blocking oligos.[139][140] The miRNA target site of an mRNA transcript can also be blocked by a steric-blocking oligo.[141] For the "in situ" detection of miRNA, LNA[142] or Morpholino[143] probes can be used. The locked conformation of LNA results in enhanced hybridization properties and increases sensitivity and selectivity, making it ideal for detection of short miRNA.[144]
High-throughput quantification of miRNAs is error prone, for the larger variance (compared to mRNAs) that comes with methodological problems. mRNA-expression is therefore often analyzed to check for miRNA-effects in their levels (e.g. in[145]). Databases can be used to pair mRNA- and miRNA-data that predict miRNA-targets based on their base sequence.[146][147] While this is usually done after miRNAs of interest have been detected (e. g. because of high expression levels), ideas for analysis tools that integrate mRNA- and miRNA-expression information have been proposed.[148][149]
Human and animal diseases
Just as miRNA is involved in the normal functioning of eukaryotic cells, so has dysregulation of miRNA been associated with disease. A manually curated, publicly available database, miR2Disease, documents known relationships between miRNA dysregulation and human disease.[150]
Inherited diseases
A mutation in the seed region of miR-96 causes hereditary progressive hearing loss.[151]
A mutation in the seed region of miR-184 causes hereditary keratoconus with anterior polar cataract.[152]
Deletion of the miR-17~92 cluster causes skeletal and growth defects.[153]
Cancer
The first human disease known to be associated with miRNA deregulation was chronic lymphocytic leukemia.[154] Many other miRNAs also have links with cancer and accordingly are sometimes referred to as "oncomirs".[155] In malignant B cells miRNAs participate in pathways fundamental to B cell development like B-cell receptor (BCR) signalling, B-cell migration/adhesion, cell-cell interactions in immune niches and the production and class-switching of immunoglobulins. MiRNAs influence B cell maturation, generation of pre-, marginal zone, follicular, B1, plasma and memory B cells.[156]
Another role for miRNA in cancers is to use their expression level for prognosis. In NSCLC samples, low miR-324a levels may serve as an indicator of poor survival.[157] Either high miR-185 or low miR-133b levels may correlate with metastasis and poor survival in colorectal cancer.[158]
Furthermore, specific miRNAs may be associated with certain histological subtypes of colorectal cancer. For instance, expression levels of miR-205 and miR-373 have been shown to be increased in mucinous colorectal cancers and mucin-producing Ulcerative Colitis-associated colon cancers, but not in sporadic colonic adenocarcinoma that lack mucinous components.[159] In-vitro studies suggested that miR-205 and miR-373 may functionally induce different features of mucinous-associated neoplastic progression in intestinal epithelial cells.[159]
Hepatocellular carcinoma cell proliferation may arise from miR-21 interaction with MAP2K3, a tumor repressor gene.[160] Optimal treatment for cancer involves accurately identifying patients for risk-stratified therapy. Those with a rapid response to initial treatment may benefit from truncated treatment regimens, showing the value of accurate disease response measures. Cell-free circulating miRNAs (cimiRNAs) are highly stable in blood, are overexpressed in cancer and are quantifiable within the diagnostic laboratory. In classical Hodgkin lymphoma, plasma miR-21, miR-494, and miR-1973 are promising disease response biomarkers.[161] Circulating miRNAs have the potential to assist clinical decision making and aid interpretation of positron emission tomography combined with computerized tomography. They can be performed at each consultation to assess disease response and detect relapse.
MicroRNAs have the potential to be used as tools or targets for treatment of different cancers.[162] The specific microRNA, miR-506 has been found to work as a tumor antagonist in several studies. A significant number of cervical cancer samples were found to have downregulated expression of miR-506. Additionally, miR-506 works to promote apoptosis of cervical cancer cells, through its direct target hedgehog pathway transcription factor, Gli3.[163][164]
DNA repair and cancer
Many miRNAs can directly target and inhibit cell cycle genes to control cell proliferation. A new strategy for tumor treatment is to inhibit tumor cell proliferation by repairing the defective miRNA pathway in tumors.[165] Cancer is caused by the accumulation of mutations from either DNA damage or uncorrected errors in DNA replication.[166] Defects in DNA repair cause the accumulation of mutations, which can lead to cancer.[167] Several genes involved in DNA repair are regulated by microRNAs.[168]
Germline mutations in DNA repair genes cause only 2–5% of colon cancer cases.[169] However, altered expression of microRNAs, causing DNA repair deficiencies, are frequently associated with cancers and may be an important causal factor. Among 68 sporadic colon cancers with reduced expression of the DNA mismatch repair protein MLH1, most were found to be deficient due to epigenetic methylation of the CpG island of the MLH1 gene.[170] However, up to 15% of MLH1-deficiencies in sporadic colon cancers appeared to be due to over-expression of the microRNA miR-155, which represses MLH1 expression.[171]
In 29–66%[172][173] of glioblastomas, DNA repair is deficient due to epigenetic methylation of the MGMT gene, which reduces protein expression of MGMT. However, for 28% of glioblastomas, the MGMT protein is deficient, but the MGMT promoter is not methylated.[172] In glioblastomas without methylated MGMT promoters, the level of microRNA miR-181d is inversely correlated with protein expression of MGMT and the direct target of miR-181d is the MGMT mRNA 3'UTR (the three prime untranslated region of MGMT mRNA).[172] Thus, in 28% of glioblastomas, increased expression of miR-181d and reduced expression of DNA repair enzyme MGMT may be a causal factor.
HMGA proteins (HMGA1a, HMGA1b and HMGA2) are implicated in cancer, and expression of these proteins is regulated by microRNAs. HMGA expression is almost undetectable in differentiated adult tissues, but is elevated in many cancers. HMGA proteins are polypeptides of ~100 amino acid residues characterized by a modular sequence organization. These proteins have three highly positively charged regions, termed AT hooks, that bind the minor groove of AT-rich DNA stretches in specific regions of DNA. Human neoplasias, including thyroid, prostatic, cervical, colorectal, pancreatic and ovarian carcinomas, show a strong increase of HMGA1a and HMGA1b proteins.[174] Transgenic mice with HMGA1 targeted to lymphoid cells develop aggressive lymphoma, showing that high HMGA1 expression is associated with cancers and that HMGA1 can act as an oncogene.[175] HMGA2 protein specifically targets the promoter of ERCC1, thus reducing expression of this DNA repair gene.[176] ERCC1 protein expression was deficient in 100% of 47 evaluated colon cancers (though the extent to which HGMA2 was involved is not known).[177]
Single Nucleotide polymorphisms (SNPs) can alter the binding of miRNAs on 3'UTRs for example the case of hsa-mir181a and hsa-mir181b on the CDON tumor suppressor gene.[178]
Heart disease
The global role of miRNA function in the heart has been addressed by conditionally inhibiting miRNA maturation in the murine heart. This revealed that miRNAs play an essential role during its development.[179][180] miRNA expression profiling studies demonstrate that expression levels of specific miRNAs change in diseased human hearts, pointing to their involvement in cardiomyopathies.[181][182][183] Furthermore, animal studies on specific miRNAs identified distinct roles for miRNAs both during heart development and under pathological conditions, including the regulation of key factors important for cardiogenesis, the hypertrophic growth response and cardiac conductance.[180][184][185][186][187][188] Another role for miRNA in cardiovascular diseases is to use their expression levels for diagnosis, prognosis or risk stratification.[189] miRNA's in animal models have also been linked to cholesterol metabolism and regulation.
miR-712
Murine microRNA-712 is a potential biomarker (i.e. predictor) for atherosclerosis, a cardiovascular disease of the arterial wall associated with lipid retention and inflammation.[190] Non-laminar blood flow also correlates with development of atherosclerosis as mechanosenors of endothelial cells respond to the shear force of disturbed flow (d-flow).[191] A number of pro-atherogenic genes including matrix metalloproteinases (MMPs) are upregulated by d-flow,[191] mediating pro-inflammatory and pro-angiogenic signals. These findings were observed in ligated carotid arteries of mice to mimic the effects of d-flow. Within 24 hours, pre-existing immature miR-712 formed mature miR-712 suggesting that miR-712 is flow-sensitive.[191] Coinciding with these results, miR-712 is also upregulated in endothelial cells exposed to naturally occurring d-flow in the greater curvature of the aortic arch.[191]
Origin
Pre-mRNA sequence of miR-712 is generated from the murine ribosomal RN45s gene at the internal transcribed spacer region 2 (ITS2).[191] XRN1 is an exonuclease that degrades the ITS2 region during processing of RN45s.[191] Reduction of XRN1 under d-flow conditions therefore leads to the accumulation of miR-712.[191]
Mechanism
MiR-712 targets tissue inhibitor of metalloproteinases 3 (TIMP3).[191] TIMPs normally regulate activity of matrix metalloproteinases (MMPs) which degrade the extracellular matrix (ECM). Arterial ECM is mainly composed of collagen and elastin fibers, providing the structural support and recoil properties of arteries.[192] These fibers play a critical role in regulation of vascular inflammation and permeability, which are important in the development of atherosclerosis.[193] Expressed by endothelial cells, TIMP3 is the only ECM-bound TIMP.[192] A decrease in TIMP3 expression results in an increase of ECM degradation in the presence of d-flow. Consistent with these findings, inhibition of pre-miR712 increases expression of TIMP3 in cells, even when exposed to turbulent flow.[191]
TIMP3 also decreases the expression of TNFα (a pro-inflammatory regulator) during turbulent flow.[191] Activity of TNFα in turbulent flow was measured by the expression of TNFα-converting enzyme (TACE) in blood. TNFα decreased if miR-712 was inhibited or TIMP3 overexpressed,[191] suggesting that miR-712 and TIMP3 regulate TACE activity in turbulent flow conditions.
Anti-miR-712 effectively suppresses d-flow-induced miR-712 expression and increases TIMP3 expression.[191] Anti-miR-712 also inhibits vascular hyperpermeability, thereby significantly reducing atherosclerosis lesion development and immune cell infiltration.[191]
Human homolog microRNA-205
The human homolog of miR-712 was found on the RN45s homolog gene, which maintains similar miRNAs to mice.[191] MiR-205 of humans share similar sequences with miR-712 of mice and is conserved across most vertebrates.[191] MiR-205 and miR-712 also share more than 50% of the cell signaling targets, including TIMP3.[191]
When tested, d-flow decreased the expression of XRN1 in humans as it did in mice endothelial cells, indicating a potentially common role of XRN1 in humans.[191]
Kidney disease
Targeted deletion of Dicer in the FoxD1-derived renal progenitor cells in a murine model resulted in a complex renal phenotype including expansion of nephron progenitors, fewer renin cells, smooth muscle arterioles, progressive mesangial loss and glomerular aneurysms.[194] High throughput whole transcriptome profiling of the FoxD1-Dicer knockout mouse model revealed ectopic upregulation of pro-apoptotic gene, Bcl2L11 (Bim) and dysregulation of the p53 pathway with increase in p53 effector genes including Bax, Trp53inp1, Jun, Cdkn1a, Mmp2, and Arid3a. p53 protein levels remained unchanged, suggesting that FoxD1 stromal miRNAs directly repress p53-effector genes. Using a lineage tracing approach followed by Fluorescent-activated cell sorting, miRNA profiling of the FoxD1-derived cells not only comprehensively defined the transcriptional landscape of miRNAs that are critical for vascular development, but also identified key miRNAs that are likely to modulate the renal phenotype in its absence. These miRNAs include miRs‐10a, 18a, 19b, 24, 30c, 92a, 106a, 130a, 152, 181a, 214, 222, 302a, 370, and 381 that regulate Bcl2L11 (Bim) and miRs‐15b, 18a, 21, 30c, 92a, 106a, 125b‐5p, 145, 214, 222, 296‐5p and 302a that regulate p53-effector genes. Consistent with the profiling results, ectopic apoptosis was observed in the cellular derivatives of the FoxD1 derived progenitor lineage and reiterates the importance of renal stromal miRNAs in cellular homeostasis.[194]
Nervous system
miRNAs appear to regulate the development and function of the nervous system.[195] Neural miRNAs are involved at various stages of synaptic development, including dendritogenesis (involving miR-132, miR-134 and miR-124), synapse formation[196] and synapse maturation (where miR-134 and miR-138 are thought to be involved).[197] Elimination of miRNA formation in mice by experimental silencing of Dicer has led to pathological outcomes, such as reduced neuronal size and motor abnormalities when silenced in striatal neurons[198] and neurodegeneration when silenced in forebrain neurons.[199] Some studies find altered miRNA expression in Alzheimer's disease,[200] as well as schizophrenia, bipolar disorder, major depression and anxiety disorders.[201][202][203]
Stroke
A Center for Disease Control and Prevention szerint a stroke az egyik vezető halál- és hosszútávúfogyaték-ok Amerikában. Az esetek 87%-a ischaemiás, mely az agyi artéria okklúziójából ered. Így az agy nem jut elég tápanyaghoz, például oxigénhez és glükózhoz, és nem távoznak onnan a fölös anyagok, például a szén-dioxid.[204][205] A miRNS-ek a cerebralis ischaemia patogenezisében lévő, például a gyulladásban, az angiogenezisben és az apoptózisban fontos gének poszttranszlációs csendesítésével működnek.[206]
Függőség
The vital role of miRNAs in gene expression is significant to addiction, specifically alcoholism.[207] Chronic alcohol abuse results in persistent changes in brain function mediated in part by alterations in gene expression.[207] miRNA global regulation of many downstream genes deems significant regarding the reorganization or synaptic connections or long term neural adaptations involving the behavioral change from alcohol consumption to withdrawal and/or dependence.[208] Up to 35 different miRNAs have been found to be altered in the alcoholic post-mortem brain, all of which target genes that include the regulation of the cell cycle, apoptosis, cell adhesion, nervous system development and cell signaling.[207] Altered miRNA levels were found in the medial prefrontal cortex of alcohol-dependent mice, suggesting the role of miRNA in orchestrating translational imbalances and the creation of differentially expressed proteins within an area of the brain where complex cognitive behavior and decision making likely originate.[209]
miRNAs can be either upregulated or downregulated in response to chronic alcohol use. miR-206 expression increased in the prefrontal cortex of alcohol-dependent rats, targeting the transcription factor brain-derived neurotrophic factor (BDNF) and ultimately reducing its expression. BDNF plays a critical role in the formation and maturation of new neurons and synapses, suggesting a possible implication in synapse growth/synaptic plasticity in alcohol abusers.[210] miR-155, important in regulating alcohol-induced neuroinflammation responses, was found to be upregulated, suggesting the role of microglia and inflammatory cytokines in alcohol pathophysiology.[211] Downregulation of miR-382 was found in the nucleus accumbens, a structure in the basal forebrain significant in regulating feelings of reward that power motivational habits. miR-382 is the target for the dopamine receptor D1 (DRD1), and its overexpression results in the upregulation of DRD1 and delta fosB, a transcription factor that activates a series of transcription events in the nucleus accumbens that ultimately result in addictive behaviors.[212] Alternatively, overexpressing miR-382 resulted in attenuated drinking and the inhibition of DRD1 and delta fosB upregulation in rat models of alcoholism, demonstrating the possibility of using miRNA-targeted pharmaceuticals in treatments.[212]
Obesitas
A miRNS-ek fontosak az őssejtprogenitorok zsírsejtté való differenciálódásának szabályzásában.[213] A pluripotens őssejtek adipogenezisben játszott szerepét a hMSC-Tert20 halhatatlanná tett csontvelő-eredetű stromalis sejtvonalban vizsgálták.[214] Decreased expression of miR-155, miR-221, and miR-222, have been found during the adipogenic programming of both immortalized and primary hMSCs, suggesting that they act as negative regulators of differentiation. Conversely, ectopic expression of the miRNAs 155, 221, and 222 significantly inhibited adipogenesis and repressed induction of the master regulators PPARγ and CCAAT/enhancer-binding protein alpha (CEBPA).[215] This paves the way for possible genetic obesity treatments.
További inzulinrezisztenciát, obesitast és diabéteszt csoportosító miRNS-osztály a let-7 család. Mennyisége az öregedés során nő.[216] A let-7 gyorsult öregedés mintájára történt ektópiás túlexpressziója egerekben inzulinrezisztenciát, így az obesitasnak és a diabétesznek való nagyobb kitettséget okozott.[217] Ezzel szemben a let-7 let-7-specifikus antagomirek injekciójával való inhibeálása nagyobb inzulinszenzitivitást és ellenállást okozott a nagyobb zsírtartalom miatti obesitasra és diabéteszre. Nemcsak gátolta az obesitas és a diabétesz kialakulását, hanem a meglévőt gyógyította.[218] Ezek alapján a let-7-gátlás új terápia lehet az obesitas és a 2-es típusú cukorbetegség ellen.
Homeosztázis
A miRNS-ek fontosak lehetnek a komplex enzimkaszkádok, például a hemosztatikus vérkoagulációs rendszer szabályzásában.[219] Funkcionális miRNS-eken végzett kísérletek 2018-ban a hemosztatikus rendszer terápiás céljait mutatták ki.[220][221] Ezek összefüggnek az endoplazmatikus retikulum kalciumhomeosztázisával, mely fontos a sejtdifferenciációhoz.[222]
Növényekben
A miRNS-ek számos fejlődési, homeosztatikus és immunfolyamat fontos szabályzói növényekben.[223] Céljuk a növényfejlődésben például a hajtások merisztémafejlődése, a levélnövekedés, a virág-, a magképzés vagy a gyökérnövekedés.[224][225][226][227] Továbbá összetett a szerepük számos hő-, alacsony hőmérsékleti, szárazság-, fénystresszel vagy γ-sugárzásnak való kitettséggel kapcsolatos abiotikus folyamatban.[223]
Vírusokban
A vírus-miRNS-ek fontosak a virális vagy gazdagének expressziójának a vírus javára való szabályzásában. Így a miRNS-ek fontosak a gazda-vírus kölcsönhatásokban és a vírusbetegségek patogenezisében.[228][229] A humán herpeszvírus-6 DNS-ének transzkripcióaktivátorainak expresszióját feltehetően vírus-miRNS-ek szabályozzák.[230]
Célelőrejelzés
A miRNS-ek fehérjekódoló gének cél-mRNS-eihez köthetnek, csökkentve transzlációjukat vagy lebontva azokat. Fontos pontos azonosításuk.[231] 18 in silico algoritmus összehasonlítása elérhető.[232] Funkcionális miRNS-célzó algoritmusok vizsgálata szerint sok funkcionális miRNS-t a célelőrejelző algoritmusok eltéveszthetnek.[220]
Jegyzetek
- ↑ a b c d Bartel DP (2018. március 1.). „Metazoan MicroRNAs”. Cell 173 (1), 20–51. o. DOI:10.1016/j.cell.2018.03.006. PMID 29570994.
- ↑ a b c Bartel DP (2004. január 1.). „MicroRNAs: genomics, biogenesis, mechanism, and function”. Cell 116 (2), 281–297. o. DOI:10.1016/S0092-8674(04)00045-5. PMID 14744438.
- ↑ Qureshi A, Thakur N, Monga I, Thakur A, Kumar M (2014. január 1.). „VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets”. Database 2014, bau103. o. DOI:10.1093/database/bau103. PMID 25380780.
- ↑ a b c Bartel DP (2009. január 1.). „MicroRNAs: target recognition and regulatory functions”. Cell 136 (2), 215–33. o. DOI:10.1016/j.cell.2009.01.002. PMID 19167326.
- ↑ a b Jonas, S (2015. július 1.). „Towards a molecular understanding of microRNA-mediated gene silencing”. Nature Reviews. Genetics 16 (7), 421–33. o. DOI:10.1038/nrg3965. PMID 26077373.
- ↑ (2010. augusztus 12.) „Mammalian microRNAs predominantly act to decrease target mRNA levels”. Nature 466 (7308), 835–40. o. DOI:10.1038/nature09267. PMID 20703300.
- ↑ 'Homo sapiens miRNAs in the miRBase. Manchesteri Egyetem
- ↑ Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, Hart M, Abu-Halima M, Grässer FA, Lenhof HP, Keller A, Meese E (2019. április 1.). „An estimate of the total number of true human miRNAs”. Nucleic Acids Research 47 (7), 3353–3364. o. DOI:10.1093/nar/gkz097. PMID 30820533.
- ↑ Fromm B, Domanska D, Høye E, Ovchinnikov V, Kang W, Aparicio-Puerta E, Johansen M, Flatmark K, Mathelier A, Hovig E, Hackenberg M, Friedländer MR, Peterson KJ (2020. január 1.). „MirGeneDB 2.0: the metazoan microRNA complement”. Nucleic Acids Research 48 (D1), D132–D141. o. DOI:10.1093/nar/gkz885. PMID 31598695.
- ↑ Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP (2003. április 1.). „The microRNAs of Caenorhabditis elegans”. Genes & Development 17 (8), 991–1008. o. DOI:10.1101/gad.1074403. PMID 12672692.
- ↑ a b Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002. április 1.). „Identification of tissue-specific microRNAs from mouse”. Current Biology 12 (9), 735–9. o. DOI:10.1016/S0960-9822(02)00809-6. PMID 12007417.
- ↑ a b c Lewis BP, Burge CB, Bartel DP (2005. január 1.). „Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets”. Cell 120 (1), 15–20. o. DOI:10.1016/j.cell.2004.12.035. PMID 15652477.
- ↑ a b c Friedman RC, Farh KK, Burge CB, Bartel DP (2009. január 1.). „Most mammalian mRNAs are conserved targets of microRNAs”. Genome Research 19 (1), 92–105. o. DOI:10.1101/gr.082701.108. PMID 18955434.
- ↑ Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ (2015). „A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome”. Annual Review of Genetics 49, 213–42. o. DOI:10.1146/annurev-genet-120213-092023. PMID 26473382.
- ↑ a b c Lee RC, Feinbaum RL, Ambros V (1993. december 1.). „The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14”. Cell 75 (5), 843–54. o. DOI:10.1016/0092-8674(93)90529-Y. PMID 8252621.
- ↑ a b Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000. február 1.). „The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans”. Nature 403 (6772), 901–6. o. DOI:10.1038/35002607. PMID 10706289.
- ↑ a b Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G (2000. november 1.). „Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA”. Nature 408 (6808), 86–9. o. DOI:10.1038/35040556. PMID 11081512.
- ↑ a b c Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001. október 1.). „Identification of novel genes coding for small expressed RNAs”. Science 294 (5543), 853–8. o. DOI:10.1126/science.1064921. PMID 11679670.
- ↑ a b c Lau NC, Lim LP, Weinstein EG, Bartel DP (2001. október 1.). „An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans”. Science 294 (5543), 858–62. o. DOI:10.1126/science.1065062. PMID 11679671.
- ↑ a b c Lee RC, Ambros V (2001. október 1.). „An extensive class of small RNAs in Caenorhabditis elegans”. Science 294 (5543), 862–4. o. DOI:10.1126/science.1065329. PMID 11679672.
- ↑ Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH (2005. július 1.). „MicroRNA expression in zebrafish embryonic development”. Science 309 (5732), 310–1. o. DOI:10.1126/science.1114519. PMID 15919954.
- ↑ a b Jones-Rhoades MW, Bartel DP, Bartel B (2006. június 27.). „MicroRNAS and their regulatory roles in plants”. Annual Review of Plant Biology 57, 19–53. o. DOI:10.1146/annurev.arplant.57.032905.105218. PMID 16669754.
- ↑ Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (2003. április 1.). „bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila”. Cell 113 (1), 25–36. o. DOI:10.1016/S0092-8674(03)00231-9. PMID 12679032.
- ↑ Cuellar TL, McManus MT (2005. december 1.). „MicroRNAs and endocrine biology”. The Journal of Endocrinology 187 (3), 327–32. o. DOI:10.1677/joe.1.06426. PMID 16423811.
- ↑ Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M (2004. november 1.). „A pancreatic islet-specific microRNA regulates insulin secretion”. Nature 432 (7014), 226–30. o. DOI:10.1038/nature03076. PMID 15538371.
- ↑ Chen CZ, Li L, Lodish HF, Bartel DP (2004. január 1.). „MicroRNAs modulate hematopoietic lineage differentiation”. Science 303 (5654), 83–6. o. DOI:10.1126/science.1091903. PMID 14657504.
- ↑ Wilfred BR, Wang WX, Nelson PT (2007. július 1.). „Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways”. Molecular Genetics and Metabolism 91 (3), 209–17. o. DOI:10.1016/j.ymgme.2007.03.011. PMID 17521938.
- ↑ Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ (2005. augusztus 1.). „The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb”. Proceedings of the National Academy of Sciences of the United States of America 102 (31), 10898–903. o. DOI:10.1073/pnas.0504834102. PMID 16040801.
- ↑ Trang P, Weidhaas JB, Slack FJ (2008. december 1.). „MicroRNAs as potential cancer therapeutics”. Oncogene 27 (Suppl 2), S52–7. o. DOI:10.1038/onc.2009.353. PMID 19956180.
- ↑ Li C, Feng Y, Coukos G, Zhang L (2009. december 1.). „Therapeutic microRNA strategies in human cancer”. The AAPS Journal 11 (4), 747–57. o. DOI:10.1208/s12248-009-9145-9. PMID 19876744.
- ↑ Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F (2010. január 1.). „microRNA: emerging therapeutic targets in acute ischemic diseases”. Pharmacology & Therapeutics 125 (1), 92–104. o. DOI:10.1016/j.pharmthera.2009.10.003. PMID 19896977.
- ↑ Hydbring P, Badalian-Very G (2013. augusztus 1.). „Clinical applications of microRNAs”. F1000Research 2, 136. o. DOI:10.12688/f1000research.2-136.v2. PMID 24627783.
- ↑ Wightman B, Ha I, Ruvkun G (1993. december 1.). „Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans”. Cell 75 (5), 855–62. o. DOI:10.1016/0092-8674(93)90530-4. PMID 8252622.
- ↑ Giza DE, Calin GA. MicroRNA and Chronic Lymphocytic Leukemia, MicroRNA: Cancer, Advances in Experimental Medicine and Biology, 23–40. o.. DOI: 10.1007/978-3-319-23730-5_2 (2015). ISBN 978-3-319-23729-9
- ↑ Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003. március 1.). „A uniform system for microRNA annotation”. RNA 9 (3), 277–9. o. DOI:10.1261/rna.2183803. PMID 12592000.
- ↑ Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006. január 1.). „miRBase: microRNA sequences, targets and gene nomenclature”. Nucleic Acids Research 34 (Database issue), D140–4. o. DOI:10.1093/nar/gkj112. PMID 16381832.
- ↑ Wright MW, Bruford EA (2011. január 1.). „Naming 'junk': human non-protein coding RNA (ncRNA) gene nomenclature”. Human Genomics 5 (2), 90–8. o. DOI:10.1186/1479-7364-5-2-90. PMID 21296742.
- ↑ Hunt M, Banerjee S, Surana P, Liu M, Fuerst G, Mathioni S, Meyers BC, Nettleton D, Wise RP (2019. június 27.). „Small RNA discovery in the interaction between barley and the powdery mildew pathogen”. BMC Genomics 20 (1), 19–53. o. DOI:10.1186/s12864-019-5947-z. PMID 31345162.
- ↑ a b c Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003. december 1.). „Prediction of mammalian microRNA targets”. Cell 115 (7), 787–98. o. DOI:10.1016/S0092-8674(03)01018-3. PMID 14697198.
- ↑ Ellwanger DC, Büttner FA, Mewes HW, Stümpflen V (2011. május 1.). „The sufficient minimal set of miRNA seed types”. Bioinformatics 27 (10), 1346–50. o. DOI:10.1093/bioinformatics/btr149. PMID 21441577.
- ↑ Rajewsky N (2006. június 1.). „microRNA target predictions in animals”. Nature Genetics 38 (6s), S8–13. o. DOI:10.1038/ng1798. PMID 16736023.
- ↑ Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005. május 1.). „Combinatorial microRNA target predictions”. Nature Genetics 37 (5), 495–500. o. DOI:10.1038/ng1536. PMID 15806104.
- ↑ Thomson DW, Bracken CP, Goodall GJ (2011. szeptember 1.). „Experimental strategies for microRNA target identification”. Nucleic Acids Research 39 (16), 6845–53. o. DOI:10.1093/nar/gkr330. PMID 21652644.
- ↑ a b Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005. február 1.). „Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs”. Nature 433 (7027), 769–73. o. DOI:10.1038/nature03315. PMID 15685193.
- ↑ Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008. szeptember 1.). „Widespread changes in protein synthesis induced by microRNAs”. Nature 455 (7209), 58–63. o. DOI:10.1038/nature07228. PMID 18668040.
- ↑ Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008. szeptember 1.). „The impact of microRNAs on protein output”. Nature 455 (7209), 64–71. o. DOI:10.1038/nature07242. PMID 18668037.
- ↑ a b Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004. október 1.). „Identification of mammalian microRNA host genes and transcription units”. Genome Research 14 (10A), 1902–10. o. DOI:10.1101/gr.2722704. PMID 15364901.
- ↑ a b c d Cai X, Hagedorn CH, Cullen BR (2004. december 1.). „Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs”. RNA 10 (12), 1957–66. o. DOI:10.1261/rna.7135204. PMID 15525708.
- ↑ Weber MJ (2005. január 1.). „New human and mouse microRNA genes found by homology search”. The FEBS Journal 272 (1), 59–73. o. DOI:10.1111/j.1432-1033.2004.04389.x. PMID 15634332.
- ↑ Kim YK, Kim VN (2007. február 1.). „Processing of intronic microRNAs”. The EMBO Journal 26 (3), 775–83. o. DOI:10.1038/sj.emboj.7601512. PMID 17255951.
- ↑ Baskerville S, Bartel DP (2005. március 1.). „Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes”. RNA 11 (3), 241–7. o. DOI:10.1261/rna.7240905. PMID 15701730.
- ↑ a b c Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004. október 1.). „MicroRNA genes are transcribed by RNA polymerase II”. The EMBO Journal 23 (20), 4051–60. o. DOI:10.1038/sj.emboj.7600385. PMID 15372072.
- ↑ Zhou X, Ruan J, Wang G, Zhang W (2007. március 1.). „Characterization and identification of microRNA core promoters in four model species”. PLOS Computational Biology 3 (3), e37. o. DOI:10.1371/journal.pcbi.0030037. PMID 17352530.
- ↑ Faller M, Guo F (2008. november 1.). „MicroRNA biogenesis: there's more than one way to skin a cat”. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1779 (11), 663–7. o. DOI:10.1016/j.bbagrm.2008.08.005. PMID 18778799.
- ↑ Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN (2003. szeptember 1.). „The nuclear RNase III Drosha initiates microRNA processing”. Nature 425 (6956), 415–9. o. DOI:10.1038/nature01957. PMID 14508493.
- ↑ Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex, MicroRNA Protocols, Methods in Molecular Biology, 33–47. o.. DOI: 10.1385/1-59745-123-1:33 (2006). ISBN 978-1-59745-123-9
- ↑ Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004. december 1.). „The Drosha-DGCR8 complex in primary microRNA processing”. Genes & Development 18 (24), 3016–27. o. DOI:10.1101/gad.1262504. PMID 15574589.
- ↑ Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN (2006. június 1.). „Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex”. Cell 125 (5), 887–901. o. DOI:10.1016/j.cell.2006.03.043. PMID 16751099.
- ↑ Conrad T, Marsico A, Gehre M, Orom UA (2014. október 1.). „Microprocessor activity controls differential miRNA biogenesisin Vivo”. Cell Reports 9 (2), 542–54. o. DOI:10.1016/j.celrep.2014.09.007. PMID 25310978.
- ↑ Auyeung VC, Ulitsky I, McGeary SE, Bartel DP (2013. február 1.). „Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing”. Cell 152 (4), 844–58. o. DOI:10.1016/j.cell.2013.01.031. PMID 23415231.
- ↑ Ali PS, Ghoshdastider U, Hoffmann J, Brutschy B, Filipek S (2012. november 1.). „Recognition of the let-7g miRNA precursor by human Lin28B”. FEBS Letters 586 (22), 3986–90. o. DOI:10.1016/j.febslet.2012.09.034. PMID 23063642.
- ↑ Ruby, JG (2007. július 5.). „Intronic microRNA precursors that bypass Drosha processing”. Nature 448 (7149), 83–6. o. DOI:10.1038/nature05983. PMID 17589500.
- ↑ (2007. július 5.) „Intronic microRNA precursors that bypass Drosha processing.”. Nature 448 (7149), 83–6. o. DOI:10.1038/nature05983. PMID 17589500.
- ↑ Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC (2007. október 1.). „Mammalian mirtron genes”. Molecular Cell 28 (2), 328–36. o. DOI:10.1016/j.molcel.2007.09.028. PMID 17964270.
- ↑ a b Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K (2008. szeptember 1.). „Frequency and fate of microRNA editing in human brain”. Nucleic Acids Research 36 (16), 5270–80. o. DOI:10.1093/nar/gkn479. PMID 18684997.
- ↑ Winter J, Jung S, Keller S, Gregory RI, Diederichs S (2009. március 1.). „Many roads to maturity: microRNA biogenesis pathways and their regulation”. Nature Cell Biology 11 (3), 228–34. o. DOI:10.1038/ncb0309-228. PMID 19255566.
- ↑ Ohman M (2007. október 1.). „A-to-I editing challenger or ally to the microRNA process”. Biochimie 89 (10), 1171–6. o. DOI:10.1016/j.biochi.2007.06.002. PMID 17628290.
- ↑ a b Murchison EP, Hannon GJ (2004. június 1.). „miRNAs on the move: miRNA biogenesis and the RNAi machinery”. Current Opinion in Cell Biology 16 (3), 223–9. o. DOI:10.1016/j.ceb.2004.04.003. PMID 15145345.

- ↑ a b c Lund E, Dahlberg JE (2006). „Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs”. Cold Spring Harbor Symposia on Quantitative Biology 71, 59–66. o. DOI:10.1101/sqb.2006.71.050. PMID 17381281.
- ↑ Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN (2011. július 1.). „Dicer recognizes the 5' end of RNA for efficient and accurate processing”. Nature 475 (7355), 201–5. o. DOI:10.1038/nature10198. PMID 21753850.
- ↑ Ji X. The Mechanism of RNase III Action: How Dicer Dices, RNA Interference, Current Topics in Microbiology and Immunology, 99–116. o.. DOI: 10.1007/978-3-540-75157-1_5 (2008). ISBN 978-3-540-75156-4
- ↑ Mirihana Arachchilage G, Dassanayake AC, Basu S (2015. február 1.). „A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation”. Chemistry & Biology 22 (2), 262–72. o. DOI:10.1016/j.chembiol.2014.12.013. PMID 25641166.
- ↑ Sohel MH (2016). „Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges”. Achievements in the Life Sciences 10 (2), 175–186. o. DOI:10.1016/j.als.2016.11.007.
- ↑ a b Boeckel JN, Reis SM, Leistner D, Thomé CE, Zeiher AM, Fichtlscherer S, Keller T (2014. április 1.). „From heart to toe: heart's contribution on peripheral microRNA levels”. International Journal of Cardiology 172 (3), 616–7. o. DOI:10.1016/j.ijcard.2014.01.082. PMID 24508494.
- ↑ Lelandais-Brière C, Sorin C, Declerck M, Benslimane A, Crespi M, Hartmann C (2010. március 1.). „Small RNA diversity in plants and its impact in development”. Current Genomics 11 (1), 14–23. o. DOI:10.2174/138920210790217918. PMID 20808519.
- ↑ Rana TM (2007. január 1.). „Illuminating the silence: understanding the structure and function of small RNAs”. Nature Reviews Molecular Cell Biology 8 (1), 23–36. o. DOI:10.1038/nrm2085. PMID 17183358.
- ↑ a b Schwarz DS, Zamore PD (2002. május 1.). „Why do miRNAs live in the miRNP?”. Genes & Development 16 (9), 1025–31. o. DOI:10.1101/gad.992502. PMID 12000786.
- ↑ Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ (2004. október 1.). „Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design”. The Journal of Biological Chemistry 279 (40), 42230–9. o. DOI:10.1074/jbc.M404931200. PMID 15292246.
- ↑ Khvorova A, Reynolds A, Jayasena SD (2003. október 1.). „Functional siRNAs and miRNAs exhibit strand bias”. Cell 115 (2), 209–16. o. DOI:10.1016/S0092-8674(03)00801-8. PMID 14567918.
- ↑ Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD (2003. október 1.). „Asymmetry in the assembly of the RNAi enzyme complex”. Cell 115 (2), 199–208. o. DOI:10.1016/S0092-8674(03)00759-1. PMID 14567917.
- ↑ Lin SL, Chang D, Ying SY (2005. augusztus 1.). „Asymmetry of intronic pre-miRNA structures in functional RISC assembly”. Gene 356, 32–8. o. DOI:10.1016/j.gene.2005.04.036. PMID 16005165.
- ↑ Okamura K, Chung WJ, Lai EC (2008. szeptember 1.). „The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs”. Cell Cycle 7 (18), 2840–5. o. DOI:10.4161/cc.7.18.6734. PMID 18769156.
- ↑ a b Pratt AJ, MacRae IJ (2009. július 1.). „The RNA-induced silencing complex: a versatile gene-silencing machine”. The Journal of Biological Chemistry 284 (27), 17897–901. o. DOI:10.1074/jbc.R900012200. PMID 19342379.
- ↑ MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA (2008. január 1.). „In vitro reconstitution of the human RISC-loading complex”. Proceedings of the National Academy of Sciences of the United States of America 105 (2), 512–7. o. DOI:10.1073/pnas.0710869105. PMID 18178619.
- ↑ Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G (2002. március 1.). „miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs”. Genes & Development 16 (6), 720–8. o. DOI:10.1101/gad.974702. PMID 11914277.
- ↑ Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T (2005. december 1.). „Identification of novel argonaute-associated proteins”. Current Biology 15 (23), 2149–55. o. DOI:10.1016/j.cub.2005.10.048. PMID 16289642.
- ↑ Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J (2005. március 1.). „Involvement of microRNA in AU-rich element-mediated mRNA instability”. Cell 120 (5), 623–34. o. DOI:10.1016/j.cell.2004.12.038. PMID 15766526.
- ↑ a b c Kai ZS, Pasquinelli AE (2010. január 1.). „MicroRNA assassins: factors that regulate the disappearance of miRNAs”. Nature Structural & Molecular Biology 17 (1), 5–10. o. DOI:10.1038/nsmb.1762. PMID 20051982.
- ↑ Chatterjee S, Grosshans H (2009. szeptember 1.). „Active turnover modulates mature microRNA activity in Caenorhabditis elegans”. Nature 461 (7263), 546–9. o. DOI:10.1038/nature08349. PMID 19734881.
- ↑ a b c Morozova N, Zinovyev A, Nonne N, Pritchard LL, Gorban AN, Harel-Bellan A (2012. szeptember 1.). „Kinetic signatures of microRNA modes of action”. RNA 18 (9), 1635–55. o. DOI:10.1261/rna.032284.112. PMID 22850425.
- ↑ Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004). „Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets”. Genome Biology 5 (9), R65. o. DOI:10.1186/gb-2004-5-9-r65. PMID 15345049.
- ↑ Kawasaki H, Taira K (2004). „MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells”. Nucleic Acids Symposium Series 48 (1), 211–2. o. DOI:10.1093/nass/48.1.211. PMID 17150553.
- ↑ a b Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008. október 1.). „Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening”. Genome Research 18 (10), 1602–9. o. DOI:10.1101/gr.080127.108. PMID 18653800.
- ↑ Mazière P, Enright AJ (2007. június 1.). „Prediction of microRNA targets”. Drug Discovery Today 12 (11–12), 452–8. o. DOI:10.1016/j.drudis.2007.04.002. PMID 17532529.
- ↑ Williams AE (2008. február 1.). „Functional aspects of animal microRNAs”. Cellular and Molecular Life Sciences 65 (4), 545–62. o. DOI:10.1007/s00018-007-7355-9. PMID 17965831.
- ↑ Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E (2009. január 1.). „Deadenylation is a widespread effect of miRNA regulation”. RNA 15 (1), 21–32. o. DOI:10.1261/rna.1399509. PMID 19029310.
- ↑ Bazzini AA, Lee MT, Giraldez AJ (2012. április 1.). „Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish”. Science 336 (6078), 233–7. o. DOI:10.1126/science.1215704. PMID 22422859.
- ↑ Djuranovic S, Nahvi A, Green R (2012. április 1.). „miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay”. Science 336 (6078), 237–40. o. DOI:10.1126/science.1215691. PMID 22499947.
- ↑ Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, Guo X, He S, Chen R (2009. február 1.). „Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells”. BMC Molecular Biology 10 (1), 12. o. DOI:10.1186/1471-2199-10-12. PMID 19232136.
- ↑ Hawkins PG, Morris KV (2008. március 1.). „RNA and transcriptional modulation of gene expression”. Cell Cycle 7 (5), 602–7. o. DOI:10.4161/cc.7.5.5522. PMID 18256543.
- ↑ Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM (2005. december 1.). „Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution”. Cell 123 (6), 1133–46. o. DOI:10.1016/j.cell.2005.11.023. PMID 16337999.
- ↑ He L, Hannon GJ (2004. július 1.). „MicroRNAs: small RNAs with a big role in gene regulation”. Nature Reviews. Genetics 5 (7), 522–531. o. DOI:10.1038/nrg1379. PMID 15211354.
- ↑ Li LC. Small RNA-Mediated Gene Activation, RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Horizon Scientific Press (2008). ISBN 978-1-904455-25-7
- ↑ Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008. február 1.). „MicroRNA-373 induces expression of genes with complementary promoter sequences”. Proceedings of the National Academy of Sciences of the United States of America 105 (5), 1608–13. o. DOI:10.1073/pnas.0707594105. PMID 18227514.
- ↑ Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011. augusztus 1.). „A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?”. Cell 146 (3), 353–8. o. DOI:10.1016/j.cell.2011.07.014. PMID 21802130.
- ↑ a b Kumar S, Reddy PH (2016. szeptember 1.). „Are circulating microRNAs peripheral biomarkers for Alzheimer's disease?”. Biochim Biophys Acta 1862 (9), 1617–27. o. DOI:10.1016/j.bbadis.2016.06.001. PMID 27264337.
- ↑ van den Berg MM, Krauskopf J, Ramaekers JG, et al. (2020. február 1.). „Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders”. Prog Neurobiol 185, 101732. o. DOI:10.1016/j.pneurobio.2019.101732. PMID 31816349.
- ↑ Cuman C, Van Sinderen M, Gantier MP, Rainczuk K, Sorby K, Rombauts L, Osianlis T, Dimitriadis E (2015. október 1.). „Human Blastocyst Secreted microRNA Regulate Endometrial Epithelial Cell Adhesion”. eBioMedicine 2 (10), 1528–1535. o. DOI:10.1016/j.ebiom.2015.09.003. PMID 26629549.
- ↑ Zhou L, Miller C, Miraglia LJ, Romero A, Mure LS, Panda S, Kay SA (2021. január 1.). „A genome-wide microRNA screen identifies the microRNA-183/96/182 cluster as a modulator of circadian rhythms”. Proceedings of the National Academy of Sciences of the United States of America 118 (1), e2020454118. o. DOI:10.1073/pnas.2020454118. PMID 33443164.
- MicroRNAs Play Key Role in Regulation of Circadian Rhythms (2021. január 6.)
- ↑ Axtell MJ, Bartel DP (2005. június 1.). „Antiquity of microRNAs and their targets in land plants”. The Plant Cell 17 (6), 1658–73. o. DOI:10.1105/tpc.105.032185. PMID 15849273.
- ↑ Tanzer A, Stadler PF (2004. május 1.). „Molecular evolution of a microRNA cluster”. Journal of Molecular Biology 339 (2), 327–35. o. DOI:10.1016/j.jmb.2004.03.065. PMID 15136036.
- ↑ Chen K, Rajewsky N (2007. február 1.). „The evolution of gene regulation by transcription factors and microRNAs”. Nature Reviews Genetics 8 (2), 93–103. o. DOI:10.1038/nrg1990. PMID 17230196.
- ↑ Lee CT, Risom T, Strauss WM (2007. április 1.). „Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny”. DNA and Cell Biology 26 (4), 209–18. o. DOI:10.1089/dna.2006.0545. PMID 17465887.
- ↑ a b c d Peterson KJ, Dietrich MR, McPeek MA (2009. július 1.). „MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion”. BioEssays 31 (7), 736–47. o. DOI:10.1002/bies.200900033. PMID 19472371.
- ↑ Shabalina SA, Koonin EV (2008. október 1.). „Origins and evolution of eukaryotic RNA interference”. Trends in Ecology & Evolution 23 (10), 578–87. o. DOI:10.1016/j.tree.2008.06.005. PMID 18715673.
- ↑ Axtell MJ, Westholm JO, Lai EC (2011. június 27.). „Vive la différence: biogenesis and evolution of microRNAs in plants and animals”. Genome Biology 12 (4), 221. o. DOI:10.1186/gb-2011-12-4-221. PMID 21554756.
- ↑ a b Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ (2009). „The deep evolution of metazoan microRNAs”. Evolution & Development 11 (1), 50–68. o. DOI:10.1111/j.1525-142X.2008.00302.x. PMID 19196333.
- ↑ Pashkovskiy PP, Ryazansky SS (2013. június 1.). „Biogenesis, evolution, and functions of plant microRNAs”. Biochemistry. Biokhimiia 78 (6), 627–37. o. DOI:10.1134/S0006297913060084. PMID 23980889.
- ↑ a b Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ (2008. február 1.). „MicroRNAs and the advent of vertebrate morphological complexity”. Proceedings of the National Academy of Sciences of the United States of America 105 (8), 2946–50. o. DOI:10.1073/pnas.0712259105. PMID 18287013.
- ↑ a b c Nozawa M, Miura S, Nei M (2010. július 1.). „Origins and evolution of microRNA genes in Drosophila species”. Genome Biology and Evolution 2, 180–9. o. DOI:10.1093/gbe/evq009. PMID 20624724.
- ↑ Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC (2004. december 1.). „Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana”. Nature Genetics 36 (12), 1282–90. o. DOI:10.1038/ng1478. PMID 15565108.
- ↑ Warthmann N, Das S, Lanz C, Weigel D (2008. május 1.). „Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae”. Molecular Biology and Evolution 25 (5), 892–902. o. DOI:10.1093/molbev/msn029. PMID 18296705.
- ↑ Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, Carrington JC (2010. április 1.). „MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana”. The Plant Cell 22 (4), 1074–89. o. DOI:10.1105/tpc.110.073999. PMID 20407027.
- ↑ Caravas J, Friedrich M (2010. június 1.). „Of mites and millipedes: recent progress in resolving the base of the arthropod tree”. BioEssays 32 (6), 488–95. o. DOI:10.1002/bies.201000005. PMID 20486135.
- ↑ Kenny NJ, Namigai EK, Marlétaz F, Hui JH, Shimeld SM (2015. december 1.). „Draft genome assemblies and predicted microRNA complements of the intertidal lophotrochozoans Patella vulgata (Mollusca, Patellogastropoda) and Spirobranchus (Pomatoceros) lamarcki (Annelida, Serpulida)”. Marine Genomics 24, 139–46. o. DOI:10.1016/j.margen.2015.07.004. PMID 26319627.
- ↑ Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, Beszteri B, Billiau K, Bonnet E, Bothwell JH, Bowler C, Boyen C, Brownlee C, Carrano CJ, Charrier B, Cho GY, Coelho SM, Collén J, Corre E, Da Silva C, Delage L, Delaroque N, Dittami SM, Doulbeau S, Elias M, Farnham G, Gachon CM, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper FC, Lang D, Le Bail A, Leblanc C, Lerouge P, Lohr M, Lopez PJ, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli CA, Nelson DR, Nyvall-Collén P, Peters AF, Pommier C, Potin P, Poulain J, Quesneville H, Read B, Rensing SA, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder DC, Ségurens B, Strittmatter M, Tonon T, Tregear JW, Valentin K, von Dassow P, Yamagishi T, Van de Peer Y, Wincker P (2010. június 1.). „The Ectocarpus genome and the independent evolution of multicellularity in brown algae”. Nature 465 (7298), 617–21. o. DOI:10.1038/nature09016. PMID 20520714.
- ↑ Cuperus JT, Fahlgren N, Carrington JC (2011. február 1.). „Evolution and functional diversification of MIRNA genes”. The Plant Cell 23 (2), 431–42. o. DOI:10.1105/tpc.110.082784. PMID 21317375.
- ↑ Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Smith SA, Putnam NH, Haddock SH, Dunn CW, Wolfsberg TG, Mullikin JC, Martindale MQ, Baxevanis AD (2013. december 1.). „The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution”. Science 342 (6164), 1242592. o. DOI:10.1126/science.1242592. PMID 24337300.
- ↑ Maxwell EK, Ryan JF, Schnitzler CE, Browne WE, Baxevanis AD (2012. december 1.). „MicroRNAs and essential components of the microRNA processing machinery are not encoded in the genome of the ctenophore Mnemiopsis leidyi”. BMC Genomics 13 (1), 714. o. DOI:10.1186/1471-2164-13-714. PMID 23256903.
- ↑ Dimond PF (2010. március 15.). „miRNAs' Therapeutic Potential”. Genetic Engineering & Biotechnology News 30 (6), 1. o. (Hozzáférés: 2010. július 10.)
- ↑ Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G (2006). „Target prediction for small, noncoding RNAs in bacteria”. Nucleic Acids Research 34 (9), 2791–802. o. DOI:10.1093/nar/gkl356. PMID 16717284.
- ↑ Liu CG, Calin GA, Volinia S, Croce CM (2008). „MicroRNA expression profiling using microarrays”. Nature Protocols 3 (4), 563–78. o. DOI:10.1038/nprot.2008.14. PMID 18388938.
- ↑ Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005. november 1.). „Real-time quantification of microRNAs by stem-loop RT-PCR”. Nucleic Acids Research 33 (20), e179. o. DOI:10.1093/nar/gni178. PMID 16314309.
- ↑ Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E (2005. szeptember 1.). „An optimized isolation and labeling platform for accurate microRNA expression profiling”. RNA 11 (9), 1461–70. o. DOI:10.1261/rna.2610405. PMID 16043497.
- ↑ Buermans HP, Ariyurek Y, van Ommen G, den Dunnen JT, 't Hoen PA (2010. december 1.). „New methods for next generation sequencing based microRNA expression profiling”. BMC Genomics 11, 716. o. DOI:10.1186/1471-2164-11-716. PMID 21171994.
- ↑ Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH (2004). „Substrate requirements for let-7 function in the developing zebrafish embryo”. Nucleic Acids Research 32 (21), 6284–91. o. DOI:10.1093/nar/gkh968. PMID 15585662.
- ↑ Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG (2007. február 1.). „Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate”. Nature Genetics 39 (2), 259–63. o. DOI:10.1038/ng1953. PMID 17220889.
- ↑ Meister G, Landthaler M, Dorsett Y, Tuschl T (2004. március 1.). „Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing”. RNA 10 (3), 544–50. o. DOI:10.1261/rna.5235104. PMID 14970398.
- ↑ Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH (2007. augusztus 1.). „Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development”. PLOS Biology 5 (8), e203. o. DOI:10.1371/journal.pbio.0050203. PMID 17676975.
- ↑ Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J (2014. január 1.). „Miravirsen (SPC3649) can inhibit the biogenesis of miR-122”. Nucleic Acids Research 42 (1), 609–21. o. DOI:10.1093/nar/gkt852. PMID 24068553.
- ↑ Choi WY, Giraldez AJ, Schier AF (2007. október 1.). „Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430”. Science 318 (5848), 271–4. o. DOI:10.1126/science.1147535. PMID 17761850.
- ↑ You Y, Moreira BG, Behlke MA, Owczarzy R (2006. május 1.). „Design of LNA probes that improve mismatch discrimination”. Nucleic Acids Research 34 (8), e60. o. DOI:10.1093/nar/gkl175. PMID 16670427.
- ↑ Lagendijk AK, Moulton JD, Bakkers J (2012. június 1.). „Revealing details: whole mount microRNA in situ hybridization protocol for zebrafish embryos and adult tissues”. Biology Open 1 (6), 566–9. o. DOI:10.1242/bio.2012810. PMID 23213449.
- ↑ Kaur H, Arora A, Wengel J, Maiti S (2006. június 1.). „Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes”. Biochemistry 45 (23), 7347–55. o. DOI:10.1021/bi060307w. PMID 16752924.
- ↑ Nielsen JA, Lau P, Maric D, Barker JL, Hudson LD (2009. augusztus 1.). „Integrating microRNA and mRNA expression profiles of neuronal progenitors to identify regulatory networks underlying the onset of cortical neurogenesis”. BMC Neuroscience 10, 98. o. DOI:10.1186/1471-2202-10-98. PMID 19689821.
- ↑ Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007. július 1.). „MicroRNA targeting specificity in mammals: determinants beyond seed pairing”. Molecular Cell 27 (1), 91–105. o. DOI:10.1016/j.molcel.2007.06.017. PMID 17612493.
- ↑ Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008. január 1.). „miRBase: tools for microRNA genomics”. Nucleic Acids Research 36 (Database issue), D154–8. o. DOI:10.1093/nar/gkm952. PMID 17991681.
- ↑ Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP (2009. július 1.). „MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression”. Nucleic Acids Research 37 (Web Server issue), W356–62. o. DOI:10.1093/nar/gkp294. PMID 19420067.
- ↑ Artmann S, Jung K, Bleckmann A, Beissbarth T (2012). „Detection of simultaneous group effects in microRNA expression and related target gene sets”. PLOS ONE 7 (6), e38365. o. DOI:10.1371/journal.pone.0038365. PMID 22723856.
- ↑ Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y (2009. január 1.). „miR2Disease: a manually curated database for microRNA deregulation in human disease”. Nucleic Acids Research 37 (Database issue), D98–104. o. DOI:10.1093/nar/gkn714. PMID 18927107.
- ↑ Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA (2009. május 1.). „Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss”. Nature Genetics 41 (5), 609–13. o. DOI:10.1038/ng.355. PMID 19363479.
- ↑ Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, Willoughby CE (2011. november 1.). „Mutation altering the miR-184 seed region causes familial keratoconus with cataract”. American Journal of Human Genetics 89 (5), 628–33. o. DOI:10.1016/j.ajhg.2011.09.014. PMID 21996275.
- ↑ de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Geneviève D, Goldenberg A, Oufadem M, Manouvrier S, Munnich A, Vidigal JA, Vekemans M, Lyonnet S, Henrion-Caude A, Ventura A, Amiel J (2011. szeptember 1.). „Germline deletion of the miR-17~92 cluster causes skeletal and growth defects in humans”. Nature Genetics 43 (10), 1026–30. o. DOI:10.1038/ng.915. PMID 21892160.
- ↑ Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell'Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM (2004. augusztus 1.). „MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias”. Proc Natl Acad Sci U S A 101 (32), 11755–60. o. DOI:10.1073/pnas.0404432101. PMID 15284443.
- ↑ (2023. január 17.) „A comprehensive investigation of histotype-specific microRNA and their variants in Stage I epithelial ovarian cancers” (angol nyelven). International Journal of Cancer 152 (9), 1989–2001. o. DOI:10.1002/ijc.34408. ISSN 0020-7136. PMID 36541726.
- ↑ Kyriakidis I, Kyriakidis K, Tsezou A (2022. augusztus 1.). „MicroRNAs and the Diagnosis of Childhood Acute Lymphoblastic Leukemia: Systematic Review, Meta-Analysis and Re-Analysis with Novel Small RNA-Seq Tools”. Cancers 14 (16), 3976. o. DOI:10.3390/cancers14163976. PMID 36010971.
- ↑ Võsa U, Vooder T, Kolde R, Fischer K, Välk K, Tõnisson N, Roosipuu R, Vilo J, Metspalu A, Annilo T (2011. október 1.). „Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer”. Genes, Chromosomes & Cancer 50 (10), 812–22. o. DOI:10.1002/gcc.20902. PMID 21748820.
- ↑ Akçakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO (2011. augusztus 1.). „miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer”. International Journal of Oncology 39 (2), 311–8. o. DOI:10.3892/ijo.2011.1043. PMID 21573504.
- ↑ a b Eyking A, Reis H, Frank M, Gerken G, Schmid KW, Cario E (2016). „MiR-205 and MiR-373 Are Associated with Aggressive Human Mucinous Colorectal Cancer”. PLOS ONE 11 (6), e0156871. o. DOI:10.1371/journal.pone.0156871. PMID 27271572.
- ↑ MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. Guangxian Xu et al., 2013
- ↑ Jones K, Nourse JP, Keane C, Bhatnagar A, Gandhi MK (2014. január 1.). „Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma”. Clinical Cancer Research 20 (1), 253–64. o. DOI:10.1158/1078-0432.CCR-13-1024. PMID 24222179.
- ↑ Hosseinahli N, Aghapour M, Duijf PH, Baradaran B (2018. augusztus 1.). „Treating cancer with microRNA replacement therapy: A literature review”. Journal of Cellular Physiology 233 (8), 5574–5588. o. DOI:10.1002/jcp.26514. PMID 29521426.
- ↑ Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang D, Parker Kerrigan BC, Shmulevich I, Chen K, Sood AK, Xue F, Zhang W (2014. július 1.). „MiR-506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer”. The Journal of Pathology 233 (3), 308–18. o. DOI:10.1002/path.4348. PMID 24604117.
- ↑ Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, Sun WW, Lou XL, Wang JH, Teng YC, Zhang ZG (2015. február 1.). „miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer”. Oncogene 34 (6), 717–25. o. DOI:10.1038/onc.2014.9. PMID 24608427.
- ↑ Peking University (2022. szeptember 13.). „Scientists Develop a New, Powerful Cancer-Fighting Weapon”. Cell 185 (11), 1888–1904.e24. o, Kiadó: SciTech Daily. DOI:10.1016/j.cell.2022.04.030. PMID 35623329. (Hozzáférés: 2022. szeptember 15.)
- ↑ Loeb KR, Loeb LA (2000. március 1.). „Significance of multiple mutations in cancer”. Carcinogenesis 21 (3), 379–385. o. DOI:10.1093/carcin/21.3.379. PMID 10688858.
- ↑ Lodish H, Berk A, Kaiser CA, Krieger M, Bretscher A, Ploegh H, Amon A, Martin KC. Molecular Cell Biology, 8th, New York: W. H. Freeman and Company, 203. o. (2016. június 27.). ISBN 978-1-4641-8339-3
- ↑ Hu H, Gatti RA (2011. június 1.). „MicroRNAs: new players in the DNA damage response”. Journal of Molecular Cell Biology 3 (3), 151–158. o. DOI:10.1093/jmcb/mjq042. PMID 21183529.
- ↑ Jasperson KW, Tuohy TM, Neklason DW, Burt RW (2010. június 1.). „Hereditary and familial colon cancer”. Gastroenterology 138 (6), 2044–58. o. DOI:10.1053/j.gastro.2010.01.054. PMID 20420945.
- ↑ Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, Bannwart F, Yurtsever H, Neuweiler J, Riehle HM, Cattaruzza MS, Heinimann K, Schär P, Jiricny J, Marra G (2005. május 1.). „Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer”. Gastroenterology 128 (5), 1160–71. o. DOI:10.1053/j.gastro.2005.01.056. PMID 15887099.
- ↑ Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, Costinean S, Sandhu SK, Nuovo GJ, Alder H, Gafa R, Calore F, Ferracin M, Lanza G, Volinia S, Negrini M, McIlhatton MA, Amadori D, Fishel R, Croce CM (2010. április 1.). „Modulation of mismatch repair and genomic stability by miR-155”. Proceedings of the National Academy of Sciences of the United States of America 107 (15), 6982–7. o. DOI:10.1073/pnas.1002472107. PMID 20351277.
- ↑ a b c Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, Jiang T, Chen CC (2012. június 1.). „miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression”. Neuro-Oncology 14 (6), 712–9. o. DOI:10.1093/neuonc/nos089. PMID 22570426.
- ↑ Spiegl-Kreinecker S, Pirker C, Filipits M, Lötsch D, Buchroithner J, Pichler J, Silye R, Weis S, Micksche M, Fischer J, Berger W (2010. január 1.). „O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients”. Neuro-Oncology 12 (1), 28–36. o. DOI:10.1093/neuonc/nop003. PMID 20150365.
- ↑ Sgarra R, Rustighi A, Tessari MA, Di Bernardo J, Altamura S, Fusco A, Manfioletti G, Giancotti V (2004. szeptember 1.). „Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer”. FEBS Letters 574 (1–3), 1–8. o. DOI:10.1016/j.febslet.2004.08.013. PMID 15358530.
- ↑ Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, Wood LJ, Huso DL, Resar LM (2004. május 1.). „The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia”. Cancer Research 64 (10), 3371–5. o. DOI:10.1158/0008-5472.CAN-04-0044. PMID 15150086.
- ↑ Borrmann L, Schwanbeck R, Heyduk T, Seebeck B, Rogalla P, Bullerdiek J, Wisniewski JR (2003. december 1.). „High mobility group A2 protein and its derivatives bind a specific region of the promoter of DNA repair gene ERCC1 and modulate its activity”. Nucleic Acids Research 31 (23), 6841–51. o. DOI:10.1093/nar/gkg884. PMID 14627817.
- ↑ Facista A, Nguyen H, Lewis C, Prasad AR, Ramsey L, Zaitlin B, Nfonsam V, Krouse RS, Bernstein H, Payne CM, Stern S, Oatman N, Banerjee B, Bernstein C (2012. április 1.). „Deficient expression of DNA repair enzymes in early progression to sporadic colon cancer”. Genome Integrity 3 (1), 3. o. DOI:10.1186/2041-9414-3-3. PMID 22494821.
- ↑ Gibert B, Delloye-Bourgeois C, Gattolliat CH, Meurette O, Le Guernevel S, Fombonne J, Ducarouge B, Lavial F, Bouhallier F, Creveaux M, Negulescu AM, Bénard J, Janoueix-Lerosey I, Harel-Bellan A, Delattre O, Mehlen P (2014. november 1.). „Regulation by miR181 family of the dependence receptor CDON tumor suppressive activity in neuroblastoma”. Journal of the National Cancer Institute 106 (11). DOI:10.1093/jnci/dju318. PMID 25313246.
- ↑ Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ (2008. február 1.). „Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure”. Proceedings of the National Academy of Sciences of the United States of America 105 (6), 2111–6. o. DOI:10.1073/pnas.0710228105. PMID 18256189.
- ↑ a b Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D (2007. április 1.). „Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2”. Cell 129 (2), 303–17. o. DOI:10.1016/j.cell.2007.03.030. PMID 17397913.
- ↑ Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J (2007. július 1.). „MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure”. Circulation 116 (3), 258–67. o. DOI:10.1161/CIRCULATIONAHA.107.687947. PMID 17606841.
- ↑ van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN (2006. november 1.). „A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure”. Proceedings of the National Academy of Sciences of the United States of America 103 (48), 18255–60. o. DOI:10.1073/pnas.0608791103. PMID 17108080.
- ↑ Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ (2007. június 1.). „Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy”. Journal of Molecular and Cellular Cardiology 42 (6), 1137–41. o. DOI:10.1016/j.yjmcc.2007.04.004. PMID 17498736.
- ↑ Zhao Y, Samal E, Srivastava D (2005. július 1.). „Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis”. Nature 436 (7048), 214–20. o. DOI:10.1038/nature03817. PMID 15951802.
- ↑ Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, Zhang Y, Yang B, Wang Z (2007. április 1.). „MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts”. The Journal of Biological Chemistry 282 (17), 12363–7. o. DOI:10.1074/jbc.C700015200. PMID 17344217.
- ↑ Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z (2007. április 1.). „The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2”. Nature Medicine 13 (4), 486–91. o. DOI:10.1038/nm1569. PMID 17401374.
- ↑ Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Høydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G (2007. május 1.). „MicroRNA-133 controls cardiac hypertrophy”. Nature Medicine 13 (5), 613–8. o. DOI:10.1038/nm1582. PMID 17468766.
- ↑ van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN (2007. április 1.). „Control of stress-dependent cardiac growth and gene expression by a microRNA”. Science 316 (5824), 575–9. o. DOI:10.1126/science.1139089. PMID 17379774.
- ↑ Keller T, Boeckel JN, Groß S, Klotsche J, Palapies L, Leistner D, Pieper L, Stalla GK, Lehnert H, Silber S, Pittrow D, Maerz W, Dörr M, Wittchen HU, Baumeister SE, Völker U, Felix SB, Dimmeler S, Zeiher AM (2017. július 1.). „Improved risk stratification in prevention by use of a panel of selected circulating microRNAs”. Scientific Reports 7 (1), 4511. o. DOI:10.1038/s41598-017-04040-w. PMID 28674420.
- ↑ Insull W (2009. január 1.). „The pathology of atherosclerosis: plaque development and plaque responses to medical treatment”. The American Journal of Medicine 122 (1 Suppl), S3–S14. o. DOI:10.1016/j.amjmed.2008.10.013. PMID 19110086.
- ↑ a b c d e f g h i j k l m n o p q Son DJ, Kumar S, Takabe W, Kim CW, Ni CW, Alberts-Grill N, Jang IH, Kim S, Kim W, Won Kang S, Baker AH, Woong Seo J, Ferrara KW, Jo H (2013). „The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis”. Nature Communications 4, 3000. o. DOI:10.1038/ncomms4000. PMID 24346612.
- ↑ a b Basu R, Fan D, Kandalam V, Lee J, Das SK, Wang X, Baldwin TA, Oudit GY, Kassiri Z (2012. december 1.). „Loss of Timp3 gene leads to abdominal aortic aneurysm formation in response to angiotensin II”. The Journal of Biological Chemistry 287 (53), 44083–96. o. DOI:10.1074/jbc.M112.425652. PMID 23144462.
- ↑ Libby P (2002). „Inflammation in atherosclerosis”. Nature 420 (6917), 868–74. o. DOI:10.1038/nature01323. PMID 12490960.
- ↑ a b Phua YL, Chu JY, Marrone AK, Bodnar AJ, Sims-Lucas S, Ho J (2015. október 1.). „Renal stromal miRNAs are required for normal nephrogenesis and glomerular mesangial survival”. Physiological Reports 3 (10), e12537. o. DOI:10.14814/phy2.12537. PMID 26438731.
- ↑ Maes OC, Chertkow HM, Wang E, Schipper HM (2009. május 1.). „MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders”. Current Genomics 10 (3), 154–68. o. DOI:10.2174/138920209788185252. PMID 19881909.
- ↑ Amin ND, Bai G, Klug JR, Bonanomi D, Pankratz MT, Gifford WD, Hinckley CA, Sternfeld MJ, Driscoll SP, Dominguez B, Lee KF, Jin X, Pfaff SL (2015. december 1.). „Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure”. Science 350 (6267), 1525–9. o. DOI:10.1126/science.aad2509. PMID 26680198.
- ↑ Schratt G (2009. december 1.). „microRNAs at the synapse”. Nature Reviews. Neuroscience 10 (12), 842–9. o. DOI:10.1038/nrn2763. PMID 19888283.
- ↑ Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT (2008. április 1.). „Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration”. Proceedings of the National Academy of Sciences 105 (14), 5614–19. o. DOI:10.1073/pnas.0801689105. PMID 18385371.
- ↑ Hébert SS, Papadopoulou AS, Smith P, Galas M, Planel E, Silahtaroglu AN, Sergeant N, Buée L, De Strooper B (2010. október 1.). „Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration”. Human Molecular Genetics 19 (20), 3959–69. o. DOI:10.1093/hmg/ddq311. PMID 20660113.
- ↑ Hosseinian S, Arefian A, Rakhsh-Khorshid H (2020. június 27.). „A meta-analysis of gene expression data highlights synaptic dysfunction in the hippocampus of brains with Alzheimer's disease”. Scientific Reports 10 (1), 8384. o. DOI:10.1038/s41598-020-64452-z. PMID 32433480.
- ↑ Hommers LG, Domschke K, Deckert J (2015. január 1.). „Heterogeneity and individuality: microRNAs in mental disorders”. Journal of Neural Transmission 122 (1), 79–97. o. DOI:10.1007/s00702-014-1338-4. PMID 25395183.
- ↑ Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS (2009. július 1.). „Evidence for X-chromosomal schizophrenia associated with microRNA alterations”. PLOS ONE 4 (7), e6121. o. DOI:10.1371/journal.pone.0006121. PMID 19568434.
- ↑ Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ (2010. december 1.). „Schizophrenia is associated with an increase in cortical microRNA biogenesis”. Molecular Psychiatry 15 (12), 1176–89. o. DOI:10.1038/mp.2009.84. PMID 19721432.
- ↑ Stroke Facts (amerikai angol nyelven). Centers for Disease Control and Prevention , 2019. március 15. (Hozzáférés: 2019. december 5.)
- ↑ Rink C, Khanna S (2011. május 1.). „MicroRNA in ischemic stroke etiology and pathology”. Physiological Genomics 43 (10), 521–528. o. DOI:10.1152/physiolgenomics.00158.2010. PMID 20841499.
- ↑ Ouyang YB, Stary CM, Yang GY, Giffard R (2013. január 1.). „microRNAs: innovative targets for cerebral ischemia and stroke”. Current Drug Targets 14 (1), 90–101. o. DOI:10.2174/138945013804806424. PMID 23170800.
- ↑ a b c Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD (2011. november 1.). „Up-regulation of microRNAs in brain of human alcoholics”. Alcoholism: Clinical and Experimental Research 35 (11), 1928–37. o. DOI:10.1111/j.1530-0277.2011.01544.x. PMID 21651580.
- ↑ Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR, Schwandt M, Sommer WH, Heilig M (2013. június 1.). „Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence”. The Pharmacogenomics Journal 13 (3), 286–96. o. DOI:10.1038/tpj.2012.17. PMID 22614244.
- ↑ Gorini G, Nunez YO, Mayfield RD (2013). „Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain”. PLOS ONE 8 (12), e82565. o. DOI:10.1371/journal.pone.0082565. PMID 24358208.
- ↑ Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A, Sun H, Schank JR, King C, Heilig M (2014. március 1.). „microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking”. The Journal of Neuroscience 34 (13), 4581–8. o. DOI:10.1523/JNEUROSCI.0445-14.2014. PMID 24672003.
- ↑ Lippai D, Bala S, Csak T, Kurt-Jones EA, Szabo G (2013). „Chronic alcohol-induced microRNA-155 contributes to neuroinflammation in a TLR4-dependent manner in mice”. PLOS ONE 8 (8), e70945. o. DOI:10.1371/journal.pone.0070945. PMID 23951048.
- ↑ a b Li J, Li J, Liu X, Qin S, Guan Y, Liu Y, Cheng Y, Chen X, Li W, Wang S, Xiong M, Kuzhikandathil EV, Ye JH, Zhang C (2013. szeptember 1.). „MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction”. EMBO Molecular Medicine 5 (9), 1402–14. o. DOI:10.1002/emmm.201201900. PMID 23873704.
- ↑ Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL (2011. szeptember 1.). „MicroRNA regulation in mammalian adipogenesis”. Experimental Biology and Medicine 236 (9), 997–1004. o. DOI:10.1258/ebm.2011.011101. PMID 21844119.
- ↑ Skårn M, Namløs HM, Noordhuis P, Wang MY, Meza-Zepeda LA, Myklebost O (2012. április 1.). „Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222”. Stem Cells and Development 21 (6), 873–83. o. DOI:10.1089/scd.2010.0503. PMID 21756067.
- ↑ Zuo Y, Qiang L, Farmer SR (2006. március 1.). „Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter”. The Journal of Biological Chemistry 281 (12), 7960–7. o. DOI:10.1074/jbc.M510682200. PMID 16431920.
- ↑ Jun-Hao ET, Gupta RR, Shyh-Chang N (2016. március 1.). „Lin28 and let-7 in the Metabolic Physiology of Aging”. Trends in Endocrinology and Metabolism 27 (3), 132–141. o. DOI:10.1016/j.tem.2015.12.006. PMID 26811207.
- ↑ Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ (2011. szeptember 1.). „The Lin28/let-7 axis regulates glucose metabolism”. Cell 147 (1), 81–94. o. DOI:10.1016/j.cell.2011.08.033. PMID 21962509.
- ↑ Frost RJ, Olson EN (2011. december 1.). „Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs”. Proceedings of the National Academy of Sciences of the United States of America 108 (52), 21075–80. o. DOI:10.1073/pnas.1118922109. PMID 22160727.
- ↑ Teruel-Montoya R, Rosendaal FR, Martínez C (2015. február 1.). „MicroRNAs in hemostasis”. Journal of Thrombosis and Haemostasis 13 (2), 170–181. o. DOI:10.1111/jth.12788. PMID 25400249.
- ↑ a b Nourse J, Braun J, Lackner K, Hüttelmaier S, Danckwardt S (2018. november 1.). „Large-scale identification of functional microRNA targeting reveals cooperative regulation of the hemostatic system”. Journal of Thrombosis and Haemostasis 16 (11), 2233–2245. o. DOI:10.1111/jth.14290. PMID 30207063.
- ↑ Nourse J, Danckwardt S (2021. február 1.). „A novel rationale for targeting FXI: Insights from the hemostatic microRNA targetome for emerging anticoagulant strategies”. Pharmacology & Therapeutics 218, 107676. o. DOI:10.1016/j.pharmthera.2020.107676. PMID 32898547.
- ↑ Berardi E, Pues M, Thorrez L, Sampaolesi M (2012. október 1.). „miRNAs in ESC differentiation”. American Journal of Physiology. Heart and Circulatory Physiology 303 (8), H931–H939. o. DOI:10.1152/ajpheart.00338.2012. PMID 22886416.
- ↑ a b Borchers A, Pieler T (2010. november 1.). „Programming pluripotent precursor cells derived from Xenopus embryos to generate specific tissues and organs”. Genes 1 (3), 413–426. o. DOI:10.3390/ijms232314755. PMID 36499082.
- ↑ Curaba J, Spriggs A, Taylor J, Li Z, Helliwell C (2012. július 1.). „miRNA regulation in the early development of barley seed”. BMC Plant Biology 12 (1), 120. o. DOI:10.1186/1471-2229-12-120. PMID 22838835.
- ↑ Choudhary A, Kumar A, Kaur H, Kaur N (2021. május 1.). „MiRNA: the taskmaster of plant world” (angol nyelven). Biologia 76 (5), 1551–1567. o. DOI:10.1007/s11756-021-00720-1. ISSN 1336-9563.
- ↑ Waheed S, Zeng L (2020. március 1.). „The Critical Role of miRNAs in Regulation of Flowering Time and Flower Development”. Genes 11 (3), 319. o. DOI:10.3390/genes11030319. PMID 32192095.
- ↑ Wong CE, Zhao YT, Wang XJ, Croft L, Wang ZH, Haerizadeh F, Mattick JS, Singh MB, Carroll BJ, Bhalla PL (2011. május 1.). „MicroRNAs in the shoot apical meristem of soybean”. Journal of Experimental Botany 62 (8), 2495–2506. o. DOI:10.1093/jxb/erq437. PMID 21504877.
- ↑ Qureshi A, Thakur N, Monga I, Thakur A, Kumar M (2014. január 1.). „VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets”. Database 2014, bau103. o. DOI:10.1093/database/bau103. PMID 25380780.
- ↑ Kumar M: VIRmiRNA. Resource for experimental viral miRNA and their targets . Bioinformatics center, CSIR-IMTECH
- ↑ Tuddenham L, Jung JS, Chane-Woon-Ming B, Dölken L, Pfeffer S (2012. február 1.). „Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B”. Journal of Virology 86 (3), 1638–49. o. DOI:10.1128/JVI.05911-11. PMID 22114334.
- ↑ Zheng H, Fu R, Wang JT, Liu Q, Chen H, Jiang SW (2013. április 1.). „Advances in the techniques for the prediction of microRNA targets”. International Journal of Molecular Sciences 14 (4), 8179–87. o. DOI:10.3390/ijms14048179. PMID 23591837.
- ↑ Agarwal V, Bell GW, Nam JW, Bartel DP (2015. augusztus 1.). „Predicting effective microRNA target sites in mammalian mRNAs”. eLife 4, e05005. o. DOI:10.7554/eLife.05005. PMID 26267216.
További információk
- miRNA definition and classification: Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003. március 1.). „A uniform system for microRNA annotation”. RNA 9 (3), 277–9. o. DOI:10.1261/rna.2183803. PMID 12592000.
- Science-összefoglaló: Baulcombe D (2002. szeptember 1.). „DNA events. An RNA microcosm”. Science 297 (5589), 2002–3. o. DOI:10.1126/science.1077906. PMID 12242426.
- miRBase adatbázis
- miRTarBase, the experimentally validated microRNA-target interactions database.
- semirna, Web application to search for microRNAs in a plant genome.
- ONCO.IO: Integrative resource for microRNA and transcription factors analysis in cancer.
- MirOB Archiválva 2014. március 4-i dátummal a Wayback Machine-ben.: MicroRNA targets database and data analysis and dataviz tool.
- ChIPBase database: An open access database for decoding the transcription factors that were involved in or affected the transcription of microRNAs from ChIP-seq data.
- An animated video of the microRNA biogenesis process.
- miRNA modulation reagents to enable up-regulation or suppression of endogenous mature microRNA function

